Rumen Fermentation—Microbiota—Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sample Collection
2.2. Determination of Rumen Fermentation Parameters
2.3. Microbial DNA Extraction and 16S rRNA Sequencing
2.4. Analysis of mRNA Levels in Rumen Epithelial Tissue
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Rumen Fermentation Parameters
3.2. Characteristics of Rumen Microflora in Different Months
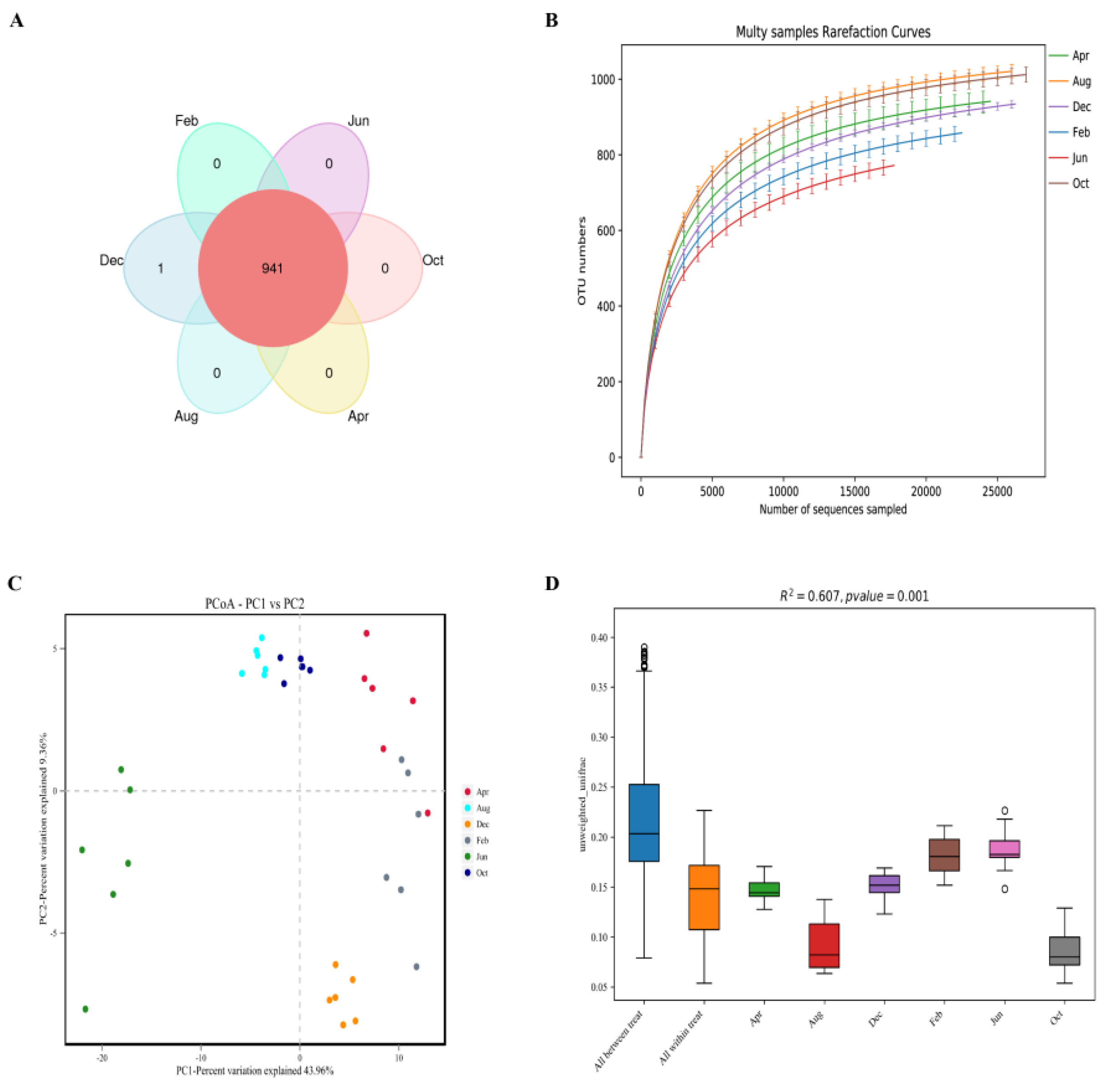
3.2.1. Rumen Microbial 16S rRNA Sequencing Results and Alpha and Beta Diversity
3.2.2. Numbers of Phyla and Genera and Differential Taxa
3.2.3. Microbe Biomarkers in Different Months
3.3. Prediction of Rumen Microbial Gene Function
3.4. Expression of Rumen Epithelial Nutrient Absorption- and Barrier-Related Genes in Different Months
3.5. Interactions among Rumen Fermentation, the Microbiota, and Host Gene Expression in Tibetan Sheep
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, P.; Gao, P.; Li, F.; Chang, S.; Wang, Z.; Yan, T.; Hou, F. Prediction of Metabolizable Energy Concentrations of Herbage in the Qinghai-Tibetan Plateau Using Tibetan Sheep Digestibility Data. Animals 2020, 10, 376. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Hu, L.; Han, X.; Zhao, N.; Xu, T.; Ma, L.; Wang, X.; Zhang, X.; Kang, S.; Zhao, X.; et al. Tibetan Sheep Adapt to Plant Phenology in Alpine Meadows by Changing Rumen Microbial Community Structure and Function. Front. Microbiol. 2020, 11, 587558. [Google Scholar] [CrossRef]
- Foster, K.R.; Schluter, J.; Coyte, K.Z.; Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 2017, 548, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Michaudel, C.; Sokol, H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 2020, 32, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Pigneur, B.; Sokol, H. Fecal microbiota transplantation in inflammatory bowel disease: The quest for the holy grail. Mucosal Immunol. 2016, 9, 1360–1365. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Wang, J.; Hou, Q.; Wang, Y.; Hu, Z.; Shi, K.; Yan, Z.; Wang, Z. Effect of hay supplementation timing on rumen microbiota in suckling calves. Microbiol. Open 2018, 7, e00430. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.E.; McEwan, N.R.; Travis, A.J.; Wallace, R.J. 16S rDNA library-based analysis of ruminal bacterial diversity. Antonie Van Leeuwenhoek 2004, 86, 263–281. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Xie, F.; Sun, D.; Liu, J.; Zhu, W.; Mao, S. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome 2019, 7, 83. [Google Scholar] [CrossRef]
- Hooper, L.V.; Midtvedt, T.; Gordon, J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 2002, 22, 283–307. [Google Scholar] [CrossRef] [Green Version]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef] [Green Version]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.Y.; Ko, H.J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef] [PubMed]
- Martin-Gallausiaux, C.; Larraufie, P.; Jarry, A.; Béguet-Crespel, F.; Marinelli, L.; Ledue, F.; Lapaque, N. Butyrate Produced by Commensal Bacteria Down-Regulates Indolamine 2,3-Dioxygenase 1 (IDO-1) Expression via a Dual Mechanism in Human Intestinal Epithelial Cells. Front. Immunol. 2018, 9, 2838. [Google Scholar] [CrossRef]
- Zhang, K.; Tu, Y.; Gao, L.; Meng, M.; Bai, Y. Replacement of grains with soybean hulls ameliorates SARA-induced impairment of the colonic epithelium barrier function of goats. BMC Vet. Res. 2018, 14, 376. [Google Scholar] [CrossRef]
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef]
- Stevenson, B.R.; Siliciano, J.D.; Mooseker, M.S.; Goodenough, D.A. Identification of ZO-1: A high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 1986, 103, 755–766. [Google Scholar] [CrossRef] [Green Version]
- Aschenbach, J.R.; Bhatia, S.K.; Pfannkuche, H.; Gäbel, G. Glucose is absorbed in a sodium-dependent manner from forestomach contents of sheep. J. Nutr. 2000, 130, 2797–2801. [Google Scholar] [CrossRef]
- Liu, X.; Sha, Y.; Dingkao, R.; Zhang, W.; Lv, W.; Wei, H.; Shi, H.; Hu, J.; Wang, J.; Li, S.; et al. Interactions Between Rumen Microbes, VFAs, and Host Genes Regulate Nutrient Absorption and Epithelial Barrier Function During Cold Season Nutritional Stress in Tibetan Sheep. Front. Microbiol. 2020, 11, 593062. [Google Scholar] [CrossRef]
- Dunphy-Doherty, F.; O’Mahony, S.M.; Peterson, V.L.; O’Sullivan, O.; Crispie, F.; Cotter, P.D.; Fone, K. Post-weaning social isolation of rats leads to long-term disruption of the gut microbiota-immune-brain axis. Brain Behav. Immun. 2018, 68, 261–273. [Google Scholar] [CrossRef]
- Miya, M.; Sato, Y.; Fukunaga, T.; Sado, T.; Poulsen, J.Y.; Sato, K.; Minamoto, T.; Yamamoto, S.; Yamanaka, H.; Araki, H. A set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2015, 2, 150088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−△△CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Liu, H.; Xu, T.; Xu, S.; Ma, L.; Han, X.; Wang, X.; Zhao, X. Effect of dietary concentrate to forage ratio on growth performance, rumen fermentation and bacterial diversity of Tibetan sheep under barn feeding on the Qinghai-Tibetan plateau. PeerJ 2019, 7, e7462. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Song, P.; Lin, G.; Huang, Y.; Wang, L.; Zhou, X.; Zhang, T. Gut Microbiota Plasticity Influences the Adaptability of Wild and Domestic Animals in Co-inhabited Areas. Front. Microbiol. 2020, 11, 125. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef]
- Long, R.J.; Apori, S.O.; Castro, F.B.; Rskov, E.R. Feed value of native forages of the tibetan plateau of china. Anim. Feed Sci. Technol. 1999, 80, 101–113. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamer, H.M.; Jonkers, D.; Venema, K.; Vanhoutvin, S.; Troost, F.J.; Brummer, R.J. Review article: The role of butyrate on colonic function. Aliment. Pharmacol. Ther. 2008, 27, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Aschenbach, J.R.; Wehning, H.; Kurze, M.; Schaberg, E.; Nieper, H.; Burckhardt, G.; Gäbel, G. Functional and molecular biological evidence of SGLT-1 in the ruminal epithelium of sheep. American journal of physiology. Gastrointest. Liver Physiol. 2000, 279, G20–G27. [Google Scholar] [CrossRef]
- Aschenbach, J.R.; Borau, T.; Gäbel, G. Glucose uptake via SGLT-1 is stimulated by beta(2)-adrenoceptors in the ruminal epithelium of sheep. J. Nutr. 2002, 132, 1254–1257. [Google Scholar] [CrossRef] [Green Version]
- Tajima, K.; Aminov, R.I.; Nagamine, T.; Matsui, H.; Nakamura, M.; Benno, Y. Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl. Environ. Microbiol. 2001, 67, 2766–2774. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Morrison, M.; Yu, Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 2011, 76, 49–63. [Google Scholar] [CrossRef] [Green Version]
- Cunha, I.S.; Barreto, C.C.; Costa, O.Y.; Bomfim, M.A.; Castro, A.P.; Kruger, R.H.; Quirino, B.F. Bacteria and Archaea community structure in the rumen microbiome of goats (Capra hircus) from the semiarid region of Brazil. Anaerobe 2011, 17, 118–124. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef]
- Shi, W.; Moon, C.D.; Leahy, S.C.; Kang, D.; Froula, J.; Kittelmann, S.; Rubin, E.M. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 2014, 24, 1517–1525. [Google Scholar] [CrossRef] [Green Version]
- Seshadri, R.; Leahy, S.C.; Attwood, G.T.; Teh, K.H.; Lambie, S.C.; Cookson, A.L.; Kelly, W.J. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat. Biotechnol. 2018, 36, 359–367. [Google Scholar] [CrossRef]
- Peng, B.; Huang, S.; Liu, T.; Geng, A. Bacterial xylose isomerases from the mammal gut Bacteroidetes cluster function in Saccharomyces cerevisiae for effective xylose fermentation. Microb. Cell Factories 2015, 14, 70. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.M.; Van Itallie, C.M. Tight junctions and the molecular basis for regulation of paracellular permeability. Am. J. Physiol. 1995, 269, G467–G475. [Google Scholar] [CrossRef]
- Yang, L.Y.; Chen, J.; Cheng, X.L.; Xi, D.M.; Yang, S.L.; Deng, W.D.; Mao, H.M. Phylogenetic analysis of 16S rRNA gene sequences reveals rumen bacterial diversity in Yaks (Bos grunniens). Mol. Biol. Rep. 2015, 37, 553–562. [Google Scholar] [CrossRef]
- Abdul Rahman, N.; Parks, D.H.; Vanwonterghem, I.; Morrison, M.; Tyson, G.W.; Hugenholtz, P. A Phylogenomic Analysis of the Bacterial Phylum Fibrobacteres. Front. Microbiol. 2016, 6, 1469. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Pitta, D.W.; Pinchak, W.E.; Indugu, N.; Vecchiarelli, B.; Sinha, R.; Fulford, J.D. Metagenomic Analysis of the Rumen Microbiome of Steers with Wheat-Induced Frothy Bloat. Front. Microbiol. 2016, 7, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H.; Global Rumen Census Collaborators. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, V.; Gupta, R.S. Molecular signatures for the phylum Synergistetes and some of its subclades. Antonie Van Leeuwenhoek 2012, 102, 517–540. [Google Scholar] [CrossRef]
- Downes, J.; Vartoukian, S.R.; Dewhirst, F.E.; Izard, J.; Chen, T.; Yu, W.H.; Wade, W.G. Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ’Synergistetes’ isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 5, 972–980. [Google Scholar] [CrossRef]
- Jumas-Bilak, E.; Roudière, L.; Marchandin, H. Description of ‘Synergistetes’ phyl. nov. and emended description of the phylum ’Deferribacteres’ and of the family Syntrophomonadaceae, phylum ’Firmicutes’. Int. J. Syst. Evol. Microbiol. 2009, 59 Pt 5, 1028–1035. [Google Scholar] [CrossRef] [Green Version]
- Khemwong, T.; Kobayashi, H.; Ikeda, Y.; Matsuura, T.; Sudo, T.; Kano, C.; Izumi, Y. Fretibacterium sp. human oral taxon 360 is a novel biomarker for periodontitis screening in the Japanese population. PLoS ONE 2019, 14, e0218266. [Google Scholar] [CrossRef] [PubMed]





| Gene | Primer (5′-3′) | Length | Annealing Temperature | Login ID |
|---|---|---|---|---|
| β-actin | F:AGCCTTCCTTCCTGGGCATGGA | 113 bp | 60 °C | NM_001009784.3 |
| R:GGACAGCACCGTGTTGGCGTAGA | ||||
| SGLT1 | F:GTGCAGTCAGCACAAAGTGG | 198 bp | 60 °C | NM_001009404.1 |
| R:CCCGGTTCCATAGGCAAACT | ||||
| CLAUDIN4 | F:AAGGTGTACGACTCGCTGCT | 237 bp | 60 °C | NM_001185017.2 |
| R:GACGTTGTTAGCCGTCCAG | ||||
| ZO1 | F:CGACCAGATCCTCAGGGTAA | 161 bp | 60 °C | XM_015101953.2 |
| R:AATCACCCACATCGGATTCT |
| Fermentation Parameters | February | April | June | August | October | December |
|---|---|---|---|---|---|---|
| Acetate (mmol/L) | 23.56 ± 0.33 d | 26.08 ± 0.81 c | 19.53 ± 1.94 e | 36.63 ± 0.33 b | 40.36 ± 0.88 a | 40.01 ± 2.01 a |
| Propionate (mmol/L) | 12.72 ± 0.61 c | 13.55 ± 1.02 bc | 10.33 ± 0.30 d | 14.37 ± 0.37 b | 15.96 ± 0.49 a | 13.86 ± 0.31 bc |
| Butyrate (mmol/L) | 6.85 ± 0.39 c | 7.48 ± 0.45 b | 5.71 ± 0.20 d | 8.11 ± 0.10 a | 8.14 ± 0.49 a | 6.05 ± 0.19 d |
| Isobutyrate (mmol/L) | 1.18 ± 0.11 a | 1.27 ± 0.09 a | 0.97 ± 0.02 b | 1.00 ± 0.01 b | 0.93 ± 0.05 bc | 0.82 ± 0.01 c |
| Isovalerate (mmol/L) | 2.53 ± 0.11 b | 2.77 ± 0.26 a | 1.85 ± 0.03 c | 1.87 ± 0.01 c | 1.69 ± 0.12 c | 1.44 ± 0.01 d |
| Valerate (mmol/L) | 5.05 ± 0.35 a | 4.61 ± 0.21 b | 4.26 ± 0.12 c | 4.13 ± 0.13 cd | 3.80 ± 0.06 d | 3.08 ± 0.15 e |
| A:P | 1.85 ± 0.06 c | 1.93 ± 0.08 c | 1.89 ± 0.13 c | 2.55 ± 0.04 b | 2.53 ± 0.02 b | 2.88 ± 0.08 a |
| Total SCFAs (mmol/L) | 51.88 ± 1.91 d | 55.77 ± 2.43 c | 42.65 ± 2.62 e | 66.11 ± 0.94 b | 70.91 ± 1.64 a | 65.27 ± 2.37 b |
| NH3-N (mg/dL) | 2.38 ± 0.03 e | 5.82 ± 0.08 c | 9.54 ± 0.26 a | 8.10 ± 0.02 b | 5.98 ± 0.03 c | 3.37 ± 0.03 d |
| CL activities (μg/min/mL) | 122.78 ± 17.25 a | 110.82 ± 15.58 ab | 110.13 ± 6.50 ab | 96.23 ± 2.01 b | 99.21 ± 4.85 b | 109.52 ± 5.58 ab |
| ACX activities (nmol/min/mL) | 144.61 ± 27.04 c | 293.32 ± 20.78 a | 257.4 ± 23.88 a | 107.64 ± 18.64 d | 162.19 ± 14.37 bc | 194.42 ± 13.55 b |
| Alpha Index | Shannon | Simpson | ACE | Chao1 | OTU | Coverage (%) |
|---|---|---|---|---|---|---|
| February | 5.24 ± 0.12 c | 0.020 ± 0.004 | 940.54 ± 13.38 c | 953.76 ± 14.19 d | 882 c | 99.60 |
| April | 5.58 ± 0.05 ab | 0.010 ± 0.001 | 1000.69 ± 24.72 b | 1015.56 ± 24.39 c | 952 b | 99.60 |
| June | 5.24 ± 0.12 c | 0.019 ± 0.007 | 891.69 ± 17.80 d | 909.56 ± 18.81 d | 819 d | 99.60 |
| August | 5.67 ± 0.08 a | 0.011 ± 0.001 | 1067.62 ± 16.55 a | 1076.28 ± 15.89 ab | 1027 a | 99.70 |
| October | 5.67 ± 0.07 a | 0.011 ± 0.002 | 1073.34 ± 14.22 a | 1092.61 ± 13.21 a | 1030 a | 99.70 |
| December | 5.35 ± 0.03 bc | 0.018 ± 0.002 | 1010.09 ± 11.01 b | 1031.25 ± 12.09 bc | 948 b | 99.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, W.; Liu, X.; Sha, Y.; Shi, H.; Wei, H.; Luo, Y.; Wang, J.; Li, S.; Hu, J.; Guo, X.; et al. Rumen Fermentation—Microbiota—Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods. Animals 2021, 11, 3529. https://doi.org/10.3390/ani11123529
Lv W, Liu X, Sha Y, Shi H, Wei H, Luo Y, Wang J, Li S, Hu J, Guo X, et al. Rumen Fermentation—Microbiota—Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods. Animals. 2021; 11(12):3529. https://doi.org/10.3390/ani11123529
Chicago/Turabian StyleLv, Weibing, Xiu Liu, Yuzhu Sha, Hao Shi, Hong Wei, Yuzhu Luo, Jiqing Wang, Shaobin Li, Jiang Hu, Xinyu Guo, and et al. 2021. "Rumen Fermentation—Microbiota—Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods" Animals 11, no. 12: 3529. https://doi.org/10.3390/ani11123529
APA StyleLv, W., Liu, X., Sha, Y., Shi, H., Wei, H., Luo, Y., Wang, J., Li, S., Hu, J., Guo, X., & Pu, X. (2021). Rumen Fermentation—Microbiota—Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods. Animals, 11(12), 3529. https://doi.org/10.3390/ani11123529






