Association of IL-4 and IL-4R Polymorphisms with Litter Size Traits in Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and DNA Extraction
2.2. Verification of Porcine IL-4 and IL-4R Polymorphisms and Genotyping
2.3. Statistical Analysis
3. Results
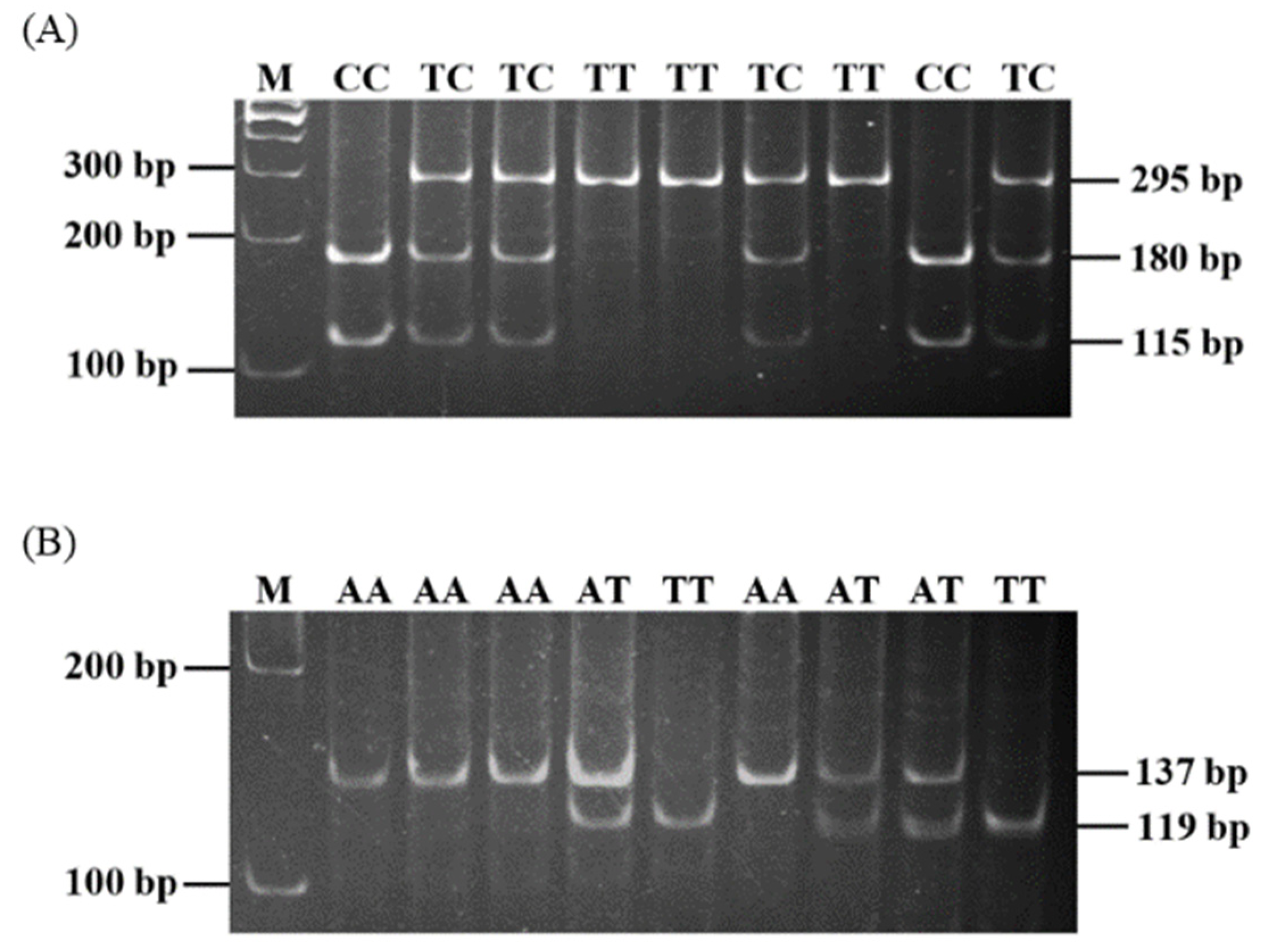
3.1. Polymorphisms of Porcine IL-4 and IL-4R Genes
3.2. Genotypic and Allelic Frequencies
3.3. Associations of Porcine IL-4 and IL-4R Polymorphisms with Litter Size Traits
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Metodev, S.; Thekkoot, D.M.; Young, J.M.; Onteru, S.; Rothschild, M.F.; Dekkers, J.C.M. A whole-genome association study for litter size and litter weight traits in pigs. Livest. Sci. 2018, 211, 87–97. [Google Scholar] [CrossRef]
- An, S.M.; Kwon, S.; Hwang, J.H.; Yu, G.E.; Kang, D.G.; Park, D.H.; Kim, T.W.; Park, H.C.; Ha, J.; Kim, C.W. Hypomethylation in the promoter region of ZPBP as a potential litter size indicator in Berkshire pigs. Arch. Anim. Breed. 2019, 62, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.F.; Rempel, L.A.; Snelling, W.M.; Weidmann, R.T.; Nonneman, D.J.; Rohrer, G.A. Genome-wide association study of swine farrowing traits. Part II: Bayesian analysis of marker data. J. Anim. Sci. 2012, 90, 3360–3367. [Google Scholar] [CrossRef] [PubMed]
- Spötter, A.; Distl, O. Genetic approaches to the improvement of fertility traits in the pig. Vet. J. 2006, 172, 234–247. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Fu, J.; Wang, A. Association of EphA4 polymorphism with swine reproductive traits and mRNA expression of EphA4 during embryo implantation. Mol. Biol. Rep. 2012, 39, 2689–2696. [Google Scholar] [CrossRef]
- Saini, V.; Arora, S.; Yadav, A.; Bhattacharjee, J. Cytokines in recurrent pregnancy loss. Clin. Chim. Acta 2011, 11, 702–708. [Google Scholar] [CrossRef]
- Geisert, R.D.; Johnson, G.A.; Burghardt, R.C. Implantation and establishment of pregnancy in the pig. Adv. Anat. Embryol. Cell Biol. 2015, 216, 137–163. [Google Scholar]
- Vélez, C.; Clauzure, M.; Williamson, D.; Koncurat, M.A.; Santa-Coloma, T.A.; Barbeito, C. IL-1β, IL-2 and IL-4 concentration during porcine gestation. Theriogenology 2019, 128, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.Y.; Ye, L.Z.; Li, F.E.; Deng, C.Y.; Jiang, S.W.; Lei, M.G.; Xiong, Y.Z. Identification of polymorphism and association analysis with reproductive traits in the porcine RNF4 gene. Anim. Reprod. Sci. 2009, 110, 283–292. [Google Scholar] [CrossRef]
- Lin, H.C.; Liu, G.F.; Wang, A.G.; Kong, L.J.; Wang, X.F.; Fu, J.L. Effect of polymorphism in the leukemia inhibitory factor gene on litter size in Large White pigs. Mol. Biol. Rep. 2009, 36, 1833–1838. [Google Scholar] [CrossRef]
- Yang, L.; Fu, J.; Fu, Y.; Wang, A. Association analysis between a polymorphism in the 5′-regulatory region of the IL-6 gene and litter size in pigs. J. Anim. Sci. Biotechnol. 2011, 2, 187–191. [Google Scholar]
- Kumchoo, T.; Mekchay, S. Association of non-synonymous SNPs of OPN gene with litter size traits in pigs. Arch. Anim. Breed. 2015, 58, 317–323. [Google Scholar] [CrossRef] [Green Version]
- Gu, T.; Zhu, M.J.; Schroyen, M.; Qu, L.; Nettleton, D.; Kuhar, D.; Lunney, J.K.; Ross, J.W.; Zhao, S.H.; Tuggle, C.K. Endometrial gene expression profiling in pregnant Meishan and Yorkshire pigs on day 12 of gestation. BMC Genom. 2014, 15, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Li, A.; Chen, W.; Wei, J.; Fu, J.; Wang, A. Differential gene expression in uterine endometrium during implantation in pigs. Biol. Reprod. 2015, 92, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, K.J.; Odukoya, O.A.; Ajjan, R.A.; Li, T.C.; Weetman, A.P.; Cooke, I.D. Profile of cytokine mRNA expression in peri-implantation human endometrium. Mol. Hum. Reprod. 1998, 4, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Ryan, J.J.; McReynolds, L.J.; Keegan, A.; Wang, L.H.; Garfein, E.; Rothman, P.; Nelms, K.; Paul, W.E. Growth and gene expression are predominantly controlled by distinct regions of the human IL-4 receptor. Immunity 1996, 4, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Arababadi, M.K.; Mosavi, R.; Ravari, A.; Teimori, H.; Hassanshahi, G. Association of interleukin-4 polymorphisms with multiple sclerosis in southeastern Iranian patients. Ann. Saudi Med. 2012, 32, 127–130. [Google Scholar] [CrossRef]
- Chatterjee, P.; Chiasson, V.L.; Bounds, K.R.; Mitchell, B.M. Regulation of the anti-inflammatory cytokines interleukin-4 and interleukin-10 during pregnancy. Front. Immunol. 2014, 27, 253. [Google Scholar] [CrossRef] [Green Version]
- Onteru, S.K.; Fan, B.; Du, Z.Q.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. A whole-genome association study for pig reproductive traits. Anim. Genet. 2012, 43, 18–26. [Google Scholar] [CrossRef]
- Schneider, J.F.; Miles, J.R.; Brown-Brandl, T.M.; Nienaber, J.A.; Rohrer, G.A.; Vallet, J.L. Genomewide association analysis for average birth interval and stillbirth in swine. J. Anim. Sci. 2015, 93, 529–540. [Google Scholar] [CrossRef]
- Guo, X.; Su, G.; Christensen, O.F.; Janss, L.; Lund, M.S. Genome-wide association analyses using a Bayesian approach for litter size and piglet mortality in Danish Landrace and Yorkshire pigs. BMC Genom. 2016, 17, 468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonneman, D.J.; Schneider, J.F.; Lents, C.A.; Wiedmann, R.T.; Vallet, J.L.; Rohrer, G.A. Genome-wide association and identification of candidate genes for age at puberty in swine. BMC Genet. 2016, 17, 50. [Google Scholar] [CrossRef] [Green Version]
- Zhu, S.; Chan-Yeung, M.; Becker, A.B.; Dimich-Ward, H.; Ferguson, A.C.; Manfreda, J.; Watson, W.T.; Paré, P.D.; Sandford, A.J. Polymorphisms of the IL-4, TNF-alpha, and Fcepsilon RIbeta genes and the risk of allergic disorders in at-risk infants. Am. J. Respir. Crit. Care Med. 2000, 161, 1655–1659. [Google Scholar] [CrossRef] [PubMed]
- Kabesch, M.; Tzotcheva, I.; Carr, D.; Höfler, C.; Weiland, S.K.; Fritzsch, C.; von Mutius, E.; Martinez, F.D. A complete screening of the IL4 gene: Novel polymorphisms and their association with asthma and IgE in childhood. J. Allergy Clin. Immunol. 2003, 112, 893–898. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, P.; Kopriva, S.E.; Chiasson, V.L.; Young, K.J.; Tobin, R.P.; Newell-Rogers, K.; Mitchell, B.M. Interleukin-4 deficiency induces mild preeclampsia in mice. J. Hypertens. 2013, 31, 1414–1423. [Google Scholar] [CrossRef]
- Salimi, S.; Khorasani, M.; Yaghmaei, M.; Mokhtari, M.; Moossavi, M. Possible association of IL-4 VNTR polymorphism with susceptibility to preeclampsia. Biomed. Res. Int. 2014, 2014, 497031. [Google Scholar] [CrossRef] [PubMed]
- Golovatyuk, K.P. Role of gene polymorphism of IL-4 and IL-17 in recurrent miscarriage, came in art cycles. Reprod. Endocrinol. 2017, 33, 26–31. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zamorano, J.; Keegan, A. A role for the insulin-interleukin (IL)-4 receptor motif of the IL-4 receptor α-chain in regulating activation of the insulin receptor substrate 2 and signal transducer and activator of transcription 6 pathways. J. Biol. Chem. 1998, 273, 9898–9905. [Google Scholar] [CrossRef] [Green Version]
- Kruse, S.; Japha, T.; Tedner, M.; Sparholt, S.H.; Forster, J.; Kuehr, J.; Deichmann, K.A. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology 1999, 96, 365–371. [Google Scholar] [CrossRef]
- Huang, J.; Liu, R.; Su, L.; Xiao, Q.; Yu, M. Transcriptome analysis revealed the embryo-induced gene expression patterns in the endometrium from Meishan and Yorkshire pigs. Int. J. Mol. Sci. 2015, 169, 22692–22710. [Google Scholar] [CrossRef] [Green Version]
- Isom, S.C.; Spollen, W.G.; Blake, S.M.; Bauer, B.K.; Springer, G.K.; Prather, R.S. Transcriptional profiling of day 12 porcine embryonic disc and trophectoderm samples using ultra-deep sequencing technologies. Mol. Reprod. Dev. 2010, 77, 812–819. [Google Scholar] [CrossRef] [PubMed]
- Verardo, L.L.; Lopes, M.S.; Mathur, P.; Madsen, O.; Silva, F.F.; Groenen, M.A.M.; Knol, E.F.; Lopes, P.S.; Guimarães, S.E.F. Gene networks for total number born in pigs across divergent environments. Mamm. Genome 2017, 28, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Goddard, K.A.; Tromp, G.; Romero, R.; Olson, J.M.; Lu, Q.; Xu, Z.; Parimi, N.; Nien, J.K.; Gomez, R.; Behnke, E.; et al. Candidate-gene association study of mothers with pre-eclampsia, and their infants, analyzing 775 SNPs in 190 genes. Hum. Hered. 2007, 63, 1–16. [Google Scholar] [CrossRef]
- Menon, R.; Pearce, B.; Velez, D.R.; Merialdi, M.; Williams, S.M.; Fortunato, S.J.; Thorsen, P. Racial disparity in pathophysiologic pathways of preterm birth based on genetic variants. Reprod. Biol. Endocrinol. 2009, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Romero, R.; Velez Edwards, D.R.; Kusanovic, J.P.; Hassan, S.S.; Mazaki-Tovi, S.; Vaisbuch, E.; Kim, C.J.; Chaiworapongsa, T.; Pearce, B.D.; Friel, L.A.; et al. Identification of fetal and maternal single nucleotide polymorphisms in candidate genes that predispose to spontaneous preterm labor with intact membranes. Am. J. Obstet. Gynecol. 2010, 202, 431. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Zeng, T.; Fan, S.; Zhang, L.; Liang, C. Genetic association between interleukin-4 receptor polymorphisms and cancer susceptibility: A meta-analysis based on 53 case-control studies. J. Cancer 2019, 26, 1538–1549. [Google Scholar] [CrossRef]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 2013, 54, 134–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muñoz, G.; Ovilo, C.; Estellé, J.; Silió, L.; Fernández, A.; Rodriguez, C. Association with litter size of new polymorphisms on ESR1 and ESR2 genes in a Chinese-European pig line. Genet. Sel. Evol. 2007, 39, 195–206. [Google Scholar] [CrossRef]
- Spencer, T.E.; Bazer, F.W. Conceptus signals for establishment and maintenance of pregnancy. Reprod. Biol. Endocrinol. 2004, 2, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross, J.W.; Ashworth, M.D.; Hurst, A.G.; Malayer, J.R.; Geisert, R.D. Analysis and characterization of differential gene expression during rapid trophoblastic elongation in the pig using suppression subtractive hybridization. Reprod. Biol. Endocrinol. 2003, 14, 23. [Google Scholar] [CrossRef]
- Bidarimath, M.; Tayade, C. Pregnancy and spontaneous fetal loss: A pig perspective. Mol. Reprod. Dev. 2017, 84, 856–869. [Google Scholar] [CrossRef] [Green Version]
- Takeda, K.; Kishimoto, T.; Akira, S. STAT6: Its role in interleukin 4-mediated biological functions. J. Mol. Med. 1997, 75, 317–326. [Google Scholar] [CrossRef]
- Bugawan, T.L.; Mirel, D.B.; Valdes, A.M.; Panelo, A.; Pozzilli, P.; Erlich, H.A. Association and interaction of the IL4R, IL4, and IL13 loci with type 1 diabetes among Filipinos. Am. J. Hum. Genet. 2003, 72, 1505–1514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghasemnejad-Berenji, H.; Ghaffari Novin, M.; Hajshafiha, M.; Nazarian, H.; Hashemi, S.M.; Ilkhanizadeh, B.; Ghasemnejad, T.; Sadeghpour, S.; Ghasemnejad-Berenji, M. Immunomodulatory effects of hydroxychloroquine on Th1/Th2 balance in women with repeated implantation failure. Biomed. Pharmacother. 2018, 107, 1277–1285. [Google Scholar] [CrossRef]
- Kwak-Kim, J.Y.; Chung-Bang, H.S.; Ng, S.C.; Ntrivalas, E.I.; Mangubat, C.P.; Beaman, K.D.; Beer, A.E.; Gilman-Sachs, A. Increased T helper 1 cytokine responses by circulating T cells are present in women with recurrent pregnancy losses and in infertile women with multiple implantation failures after IVF. Hum. Reprod. 2003, 18, 767–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faffe, D.S.; Flynt, L.; Bourgeois, K.; Panettieri, R.A., Jr.; Shore, S.A. Interleukin-13 and interleukin-4 induce vascular endothelial growth factor release from airway smooth muscle cells: Role of vascular endothelial growth factor genotype. Am. J. Respir. Cell Mol. Biol. 2006, 34, 213–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccinni, M.P.; Beloni, L.; Livi, C.; Maggi, E.; Scarselli, G.; Romagnani, S. Defective production of both leukemia inhibitory factor and type 2 T-helper cytokines by decidual T cells in unexplained recurrent abortions. Nat. Med. 1998, 49, 1020–1024. [Google Scholar] [CrossRef] [PubMed]
- Saleh, A.; Stathopoulou, M.G.; Dadé, S.; Ndiaye, N.C.; Azimi-Nezhad, M.; Murray, H.; Masson, C.; Lamont, J.; Fitzgerald, P.; Visvikis-Siest, S. Angiogenesis related genes NOS3, CD14, MMP3 and IL4R are associated to VEGF gene expression and circulating levels in healthy adults. BMC Med. Genet. 2015, 16, 90. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, P.; Jana, S.K.; Pasricha, P.; Ghosh, S.; Chakravarty, B.; Chaudhury, K. Proinflammatory cytokines induced altered expression of cyclooxygenase-2 gene results in unreceptive endometrium in women with idiopathic recurrent spontaneous miscarriage. Fertil. Steril. 2013, 99, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.J.; McReynolds, L.J.; Huang, H.; Nelms, K.; Paul, W.E. Characterization of a mobile Stat6 activation motif in the human IL-4 receptor. J. Immunol. 1998, 161, 1811–1821. [Google Scholar]
- Taylor, B.D.; Tang, G.; Ness, R.B.; Olsen, J.; Hougaard, D.M.; Skogstrand, K.; Roberts, J.M.; Haggerty, C.L. Mid-pregnancy circulating immune biomarkers in women with preeclampsia and normotensive controls. Pregnancy Hypertens. 2016, 6, 72–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tavasolian, F.; Abdollahi, E.; Samadi, M. Association of the IL4R single-nucleotide polymorphism I50V with recurrent spontaneous abortion (RSA). J. Assist. Reprod. Genet. 2014, 31, 851–856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Servin, B.; Martin, O.C.; Mézard, M.; Hospital, F. Toward a theory of marker-assisted gene pyramiding. Genetics 2004, 168, 513–523. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, A.; Fu, J.; Lin, H. Effects of ESR1, FSHB and RBP4 genes on litter size in a Large White and Landrace herds. Arch. Anim. Breed. 2006, 49, 64–70. [Google Scholar] [CrossRef] [Green Version]
- Noguera, J.L.; Rodríguez, C.; Varona, L.; Tomàs, A.; Muñoz, G.; Ramírez, O.; Barragán, C.; Arqué, M.; Bidanel, J.P.; Amills, M.; et al. A bi-dimensional genome scan for prolificacy traits in pigs shows the existence of multiple epistatic QTL. BMC Genom. 2009, 10, 636. [Google Scholar] [CrossRef]
- Landi, S.; Bottari, F.; Gemignani, F.; Gioia-Patricola, L.; Guino, E.; Osorio, A.; de Oca, J.; Capella, G.; Canzian, F.; Moreno, V.; et al. Interleukin-4 and interleukin-4 receptor polymorphisms and colorectal cancer risk. Eur. J. Cancer 2007, 43, 762–768. [Google Scholar] [CrossRef]
- Toumi, A.; Abida, O.; Ben Ayed, M.; Masmoudi, A.; Turki, H.; Masmoudi, H. Cytokine gene polymorphisms in Tunisian endemic pemphigus foliaceus: A possible role of il-4 variants. Hum. Immunol. 2003, 74, 658–665. [Google Scholar] [CrossRef]

| Primers | Location | Primer Sequences | PCR Size (bp) | Ta (°C) |
|---|---|---|---|---|
| IL-4-1 | Exon 1 | F: 5′-GTTAGCTCCTCCCAGTAAAC-3′ R: 5′-CATCAGAGCATCTGGAGAGA-3′ | 236 | 60 |
| IL-4-2 | Exon 2 | F: 5′-AACGCTCTCTCTTGCTCTCT-3′ R: 5′-TGCCTCCTGTATTCCAGGTA-3′ | 409 | 60 |
| IL-4-3 | Exon 3 | F: 5′-ACACGGTAATTGGTGGCTCT-3′ R: 5′-TGTTAGAAGCCAAGCTGTGG-3′ | 189 | 60 |
| IL-4-4 | Exon 4 | F: 5′-AGGGAGAAAGGCATGAGCAA-3′ R: 5′-GCCTATGCCATTAAAGTGACA-3′ | 227 | 60 |
| IL-4-5 | Intron 2 | F: 5′-GTACACGCCCTCTTCTAAGA-3′ R: 5′-TCCCTGGAAATCTCACATGC-3′ | 308 | 60 |
| IL-4-6 | Intron 3 | F: 5′-GTGGTAGGTATCCTTTCCAC-3′ R: 5′-AACAGTGATCAAACCAGGGC-3′ | 333 | 58 |
| SNP Position (SNP ID) | Location | Primer Sequences | PCR Size (bp) | Ta (°C) | Restriction Enzyme |
|---|---|---|---|---|---|
| IL-4 g.134993898T > C (rs329453960) | Intron 3 | F: 5′-GTGGTAGGTATCCTTTCCAC-3′ R: 5′-AACAGTGATCAAACCAGGGC-3′ | 333 | 58 | BsuRI |
| IL-4R c.163G > A (rs342744061) | Exon 1 | F: 5′-TCTTGATCACTGGGCTTCCG-3′ R: 5′-AACTCAGCGCTGCAGTTGAC-3′ | 152 | 60 | PflFI |
| IL-4R c.242C > T (rs334778260) | Exon 2 | F: 5′-GGTCACATGACCAGCCTAAT-3′ R: 5′-TTGAAGGAGCTGTTCCACAG-3′ | 180 | 60 | AflIII |
| IL-4R c.623G > A (rs790596006) | Exon 4 | F: 5′-TCTATAACGTGACCTACCTG-3′ R: 5′-TTAAGCCACTTGACACTCGG-3′ | 148 | 60 | MluI |
| IL-4R c.1016G > T (rs692527061) | Exon 8 | F: 5′-GCTGGAAGACTTGTCTTACC-3′ R: 5′-GATCGTCTTGCTGACCTCTA-3′ | 149 | 60 | BsuRI |
| IL-4R c.1577A > T (rs342791614) | Exon 8 | F: 5′-CTGGACTCGGACCCAGAG-3′ R: 5′-ACACTCTGGCGCAGGATCT-3′ | 137 | 58 | AluI |
| SNPs | n | Genotypic Frequencies | Allelic Frequencies 1 | p-Value 2 (χ2) | |||
|---|---|---|---|---|---|---|---|
| AA | AB | BB | A | B | |||
| IL-4 g.134993898T > C | 320 | 0.41 | 0.47 | 0.12 | 0.65 | 0.35 | 0.90 |
| IL-4R c.1577A > T | 318 | 0.28 | 0.55 | 0.17 | 0.56 | 0.44 | 0.10 |
| Parity | Traits 1 | Genotypes (Means ± SE) 2 | Additive | Dominance | ||
|---|---|---|---|---|---|---|
| TT | TC | CC | ||||
| First parity | n | 132 | 150 | 38 | ||
| TNB | 9.58 ± 0.45 | 9.77 ± 0.48 | 9.68 ± 0.67 | −0.05 ± 0.25 | 0.14 ± 0.32 | |
| NBA | 8.65 ± 0.47 | 8.57 ± 0.36 | 8.08 ± 0.42 | 0.29 ± 0.27 | 0.11 ± 0.12 | |
| NWA | 8.15 ± 0.47 | 7.85 ± 0.64 | 7.28 ± 0.54 | 0.44 ± 0.22 | 0.14 ± 0.12 | |
| MBW | 1.58 ± 0.06 | 1.60 ± 0.08 | 1.67 ± 0.08 | −0.04 ± 0.02 | −0.02 ± 0.02 | |
| MWW | 6.42 ± 0.12 | 6.58 ± 0.15 | 6.52 ± 0.18 | −0.05 ± 0.05 | 0.11 ± 0.08 | |
| Later parities (2nd–8th parities) | n | 348 | 380 | 97 | ||
| TNB | 11.25 ± 0.51 | 11.03 ± 0.53 | 10.72 ± 0.55 | 0.27 ± 0.28 | 0.04 ± 0.17 | |
| NBA | 10.48 ± 0.58 | 9.92 ± 0.57 | 9.47 ± 0.75 | 0.51 ± 0.32 | 0.06 ± 0.32 | |
| NWA | 9.72 ± 0.47 b | 9.42 ± 0.47 b | 8.45 ± 0.52 a | 0.64 ± 0.21 * | 0.33 ± 0.34 | |
| MBW | 1.58 ± 0.05 | 1.57 ± 0.04 | 1.54 ± 0.05 | 0.02 ± 0.01 | 0.01 ± 0.02 | |
| MWW | 6.62 ± 0.05 | 6.67 ± 0.05 | 6.59 ± 0.07 | 0.02 ± 0.02 | 0.06 ± 0.02 | |
| Parity | Traits 1 | Genotypes (Means ± SE) 2 | Additive | Dominance | ||
|---|---|---|---|---|---|---|
| AA | AT | TT | ||||
| First parity | n | 89 | 176 | 53 | ||
| TNB | 9.48 ± 0.38 | 10.03 ± 0.76 | 9.27 ± 0.58 | 0.11 ± 0.32 | 0.65 ± 0.35 | |
| NBA | 8.25 ± 0.37 | 8.55 ± 0.52 | 8.19 ± 0.67 | 0.03 ± 0.32 | 0.33 ± 0.47 | |
| NWA | 7.45 ± 0.53 | 7.57 ± 0.61 | 7.30 ± 0.49 | 0.08 ± 0.35 | 0.20 ± 0.42 | |
| MBW | 1.54 ± 0.04 | 1.58 ± 0.08 | 1.67 ± 0.08 | −0.06 ± 0.02 | −0.02 ± 0.03 | |
| MWW | 6.42 ± 0.12 | 6.39 ± 0.10 | 6.55 ± 0.18 | −0.06 ± 0.05 | −0.09 ± 0.07 | |
| Later parities (2nd–8th parities) | n | 235 | 449 | 133 | ||
| TNB | 10.51 ± 0.49 | 10.55 ± 0.43 | 11.54 ± 0.68 | −0.52 ± 0.28 | −0.47 ± 0.32 | |
| NBA | 9.21 ± 0.56 a | 9.43 ± 0.47 a | 10.71 ± 0.64 b | −0.75 ± 0.28 * | −0.53 ± 0.36 | |
| NWA | 8.61 ± 0.52 a | 9.09 ± 0.46 a | 10.20 ± 0.58 b | −0.80 ± 0.28 ** | −0.32 ± 0.31 | |
| MBW | 1.55 ± 0.05 | 1.60 ± 0.04 | 1.57 ± 0.05 | −0.01 ± 0.02 | 0.04 ± 0.02 | |
| MWW | 6.62 ± 0.06 | 6.63 ± 0.05 | 6.60 ± 0.06 | 0.01 ± 0.02 | 0.02 ± 0.03 | |
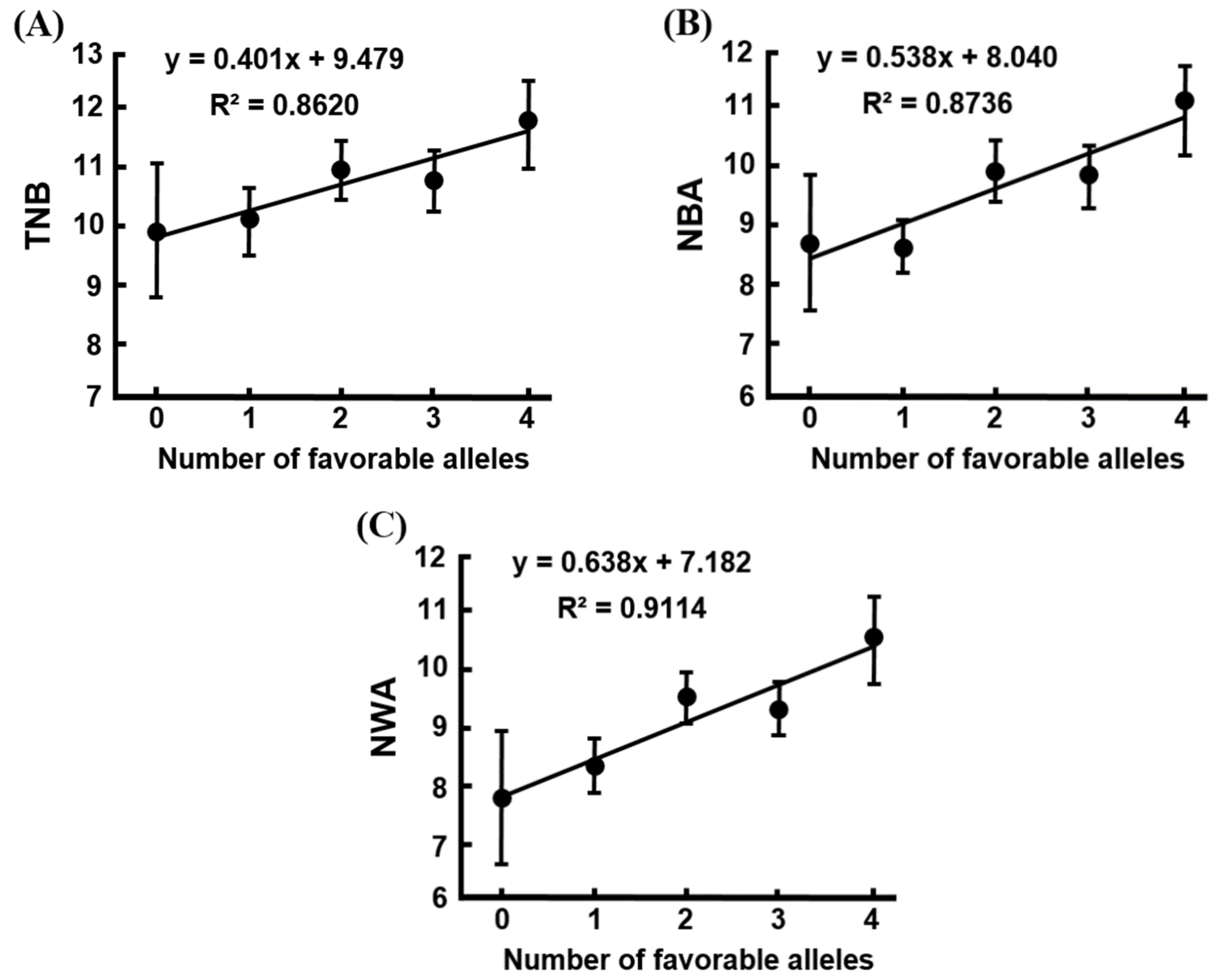
| Parity | Traits 1 | Number of Favorable Alleles (Means ± SE) 2 | ||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | ||
| First parity | n | 7 | 59 | 144 | 89 | 18 |
| TNB | 10.72 ± 1.42 | 9.58 ± 0.57 | 10.12 ± 0.54 | 10.15 ± 0.52 | 8.68 ± 0.72 | |
| NBA | 8.84 ± 1.25 | 8.24 ± 0.58 | 8.74 ± 0.52 | 9.15 ± 0.67 | 8.12 ± 0.95 | |
| NWA | 7.23 ± 1.27 | 7.25 ± 0.73 | 7.88 ± 0.64 | 8.57 ± 0.71 | 7.82 ± 0.84 | |
| MBW | 1.48 ± 0.16 | 1.58 ± 0.05 | 1.57 ± 0.04 | 1.56 ± 0.05 | 1.58 ± 0.08 | |
| MWW | 6.52 ± 0.32 | 6.41 ± 0.12 | 6.55 ± 0.10 | 6.43 ± 0.08 | 6.64 ± 0.19 | |
| Later parities (2nd–8th parities) | n | 18 | 152 | 371 | 225 | 48 |
| TNB | 9.94 ± 1.14 | 10.21 ± 0.62 | 10.87 ± 0.51 | 10.68 ± 0.43 | 11.71 ± 0.78 | |
| NBA | 8.81 ± 1.17 ab | 8.75 ± 0.55 a | 9.91 ± 0.46 b | 9.85 ± 0.47 b | 10.95 ± 0.77 b | |
| NWA | 7.81 ± 1.11 ab | 8.34 ± 0.55 a | 9.51 ± 0.43 bc | 9.30 ± 0.47 bc | 10.52 ± 0.72 c | |
| MBW | 1.65 ± 0.07 | 1.61 ± 0.05 | 1.63 ± 0.04 | 1.66 ± 0.05 | 1.58 ± 0.07 | |
| MWW | 6.21 ± 0.11 a | 6.64 ± 0.04 b | 6.65 ± 0.03 b | 6.63 ± 0.03 b | 6.65 ± 0.05 b | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norseeda, W.; Liu, G.; Teltathum, T.; Supakankul, P.; Sringarm, K.; Naraballobh, W.; Khamlor, T.; Chomdej, S.; Nganvongpanit, K.; Krutmuang, P.; et al. Association of IL-4 and IL-4R Polymorphisms with Litter Size Traits in Pigs. Animals 2021, 11, 1154. https://doi.org/10.3390/ani11041154
Norseeda W, Liu G, Teltathum T, Supakankul P, Sringarm K, Naraballobh W, Khamlor T, Chomdej S, Nganvongpanit K, Krutmuang P, et al. Association of IL-4 and IL-4R Polymorphisms with Litter Size Traits in Pigs. Animals. 2021; 11(4):1154. https://doi.org/10.3390/ani11041154
Chicago/Turabian StyleNorseeda, Worrarak, Guisheng Liu, Tawatchai Teltathum, Pantaporn Supakankul, Korawan Sringarm, Watcharapong Naraballobh, Trisadee Khamlor, Siriwadee Chomdej, Korakot Nganvongpanit, Patcharin Krutmuang, and et al. 2021. "Association of IL-4 and IL-4R Polymorphisms with Litter Size Traits in Pigs" Animals 11, no. 4: 1154. https://doi.org/10.3390/ani11041154









