Effects of Incorporating Dry Matter Intake and Residual Feed Intake into a Selection Index for Dairy Cattle Using Deterministic Modeling
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.1.1. Trait Definitions
2.1.2. Data
2.2. Genetic Parameter Estimation
2.3. Modeling Software
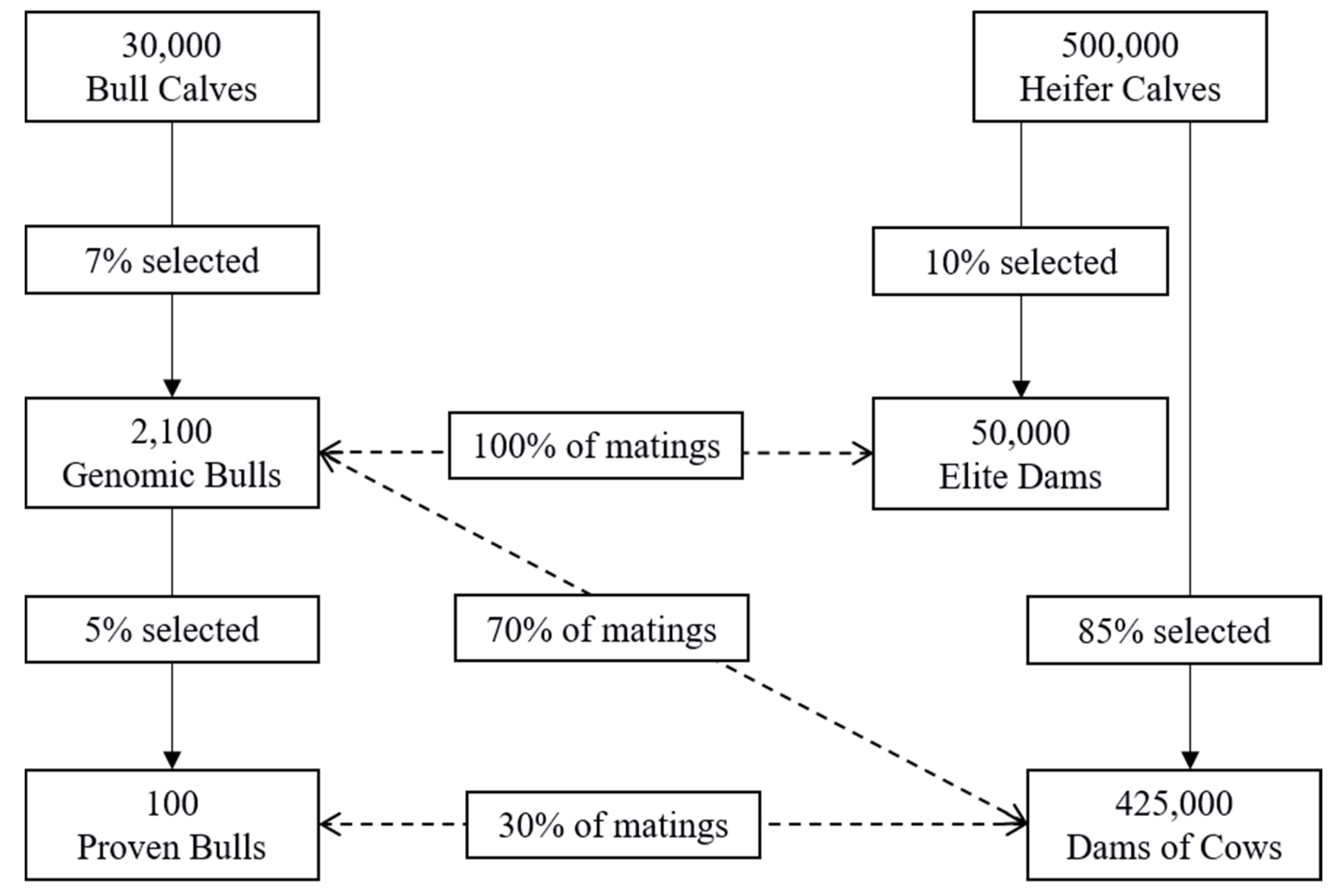
2.4. Population Structure
2.5. Selection Scenarios
2.6. Economic Values
3. Results
3.1. Genetic Parameters
3.2. Direct vs. Indirect Selection on Feed Efficiency
3.2.1. Index Response
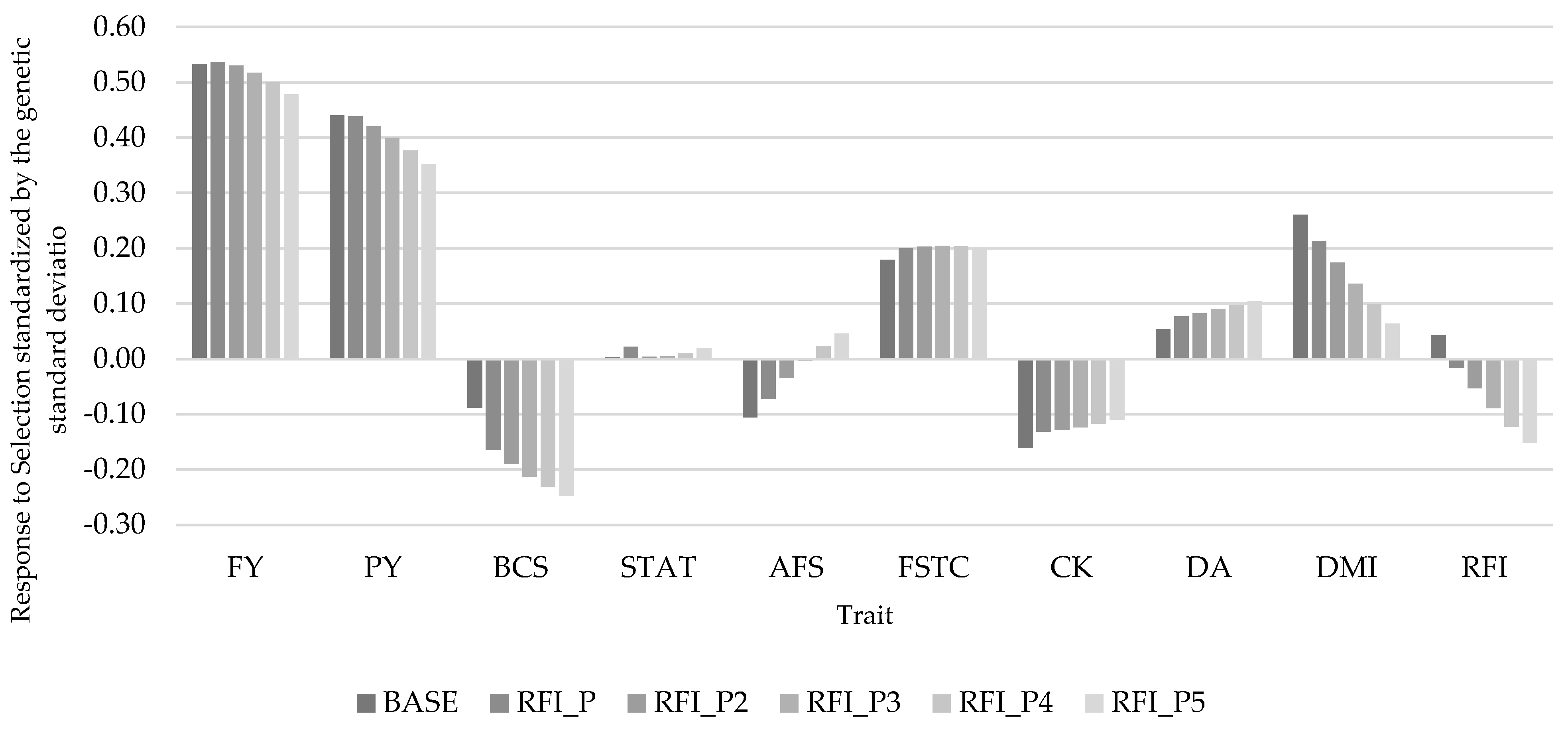
3.2.2. Trait Response
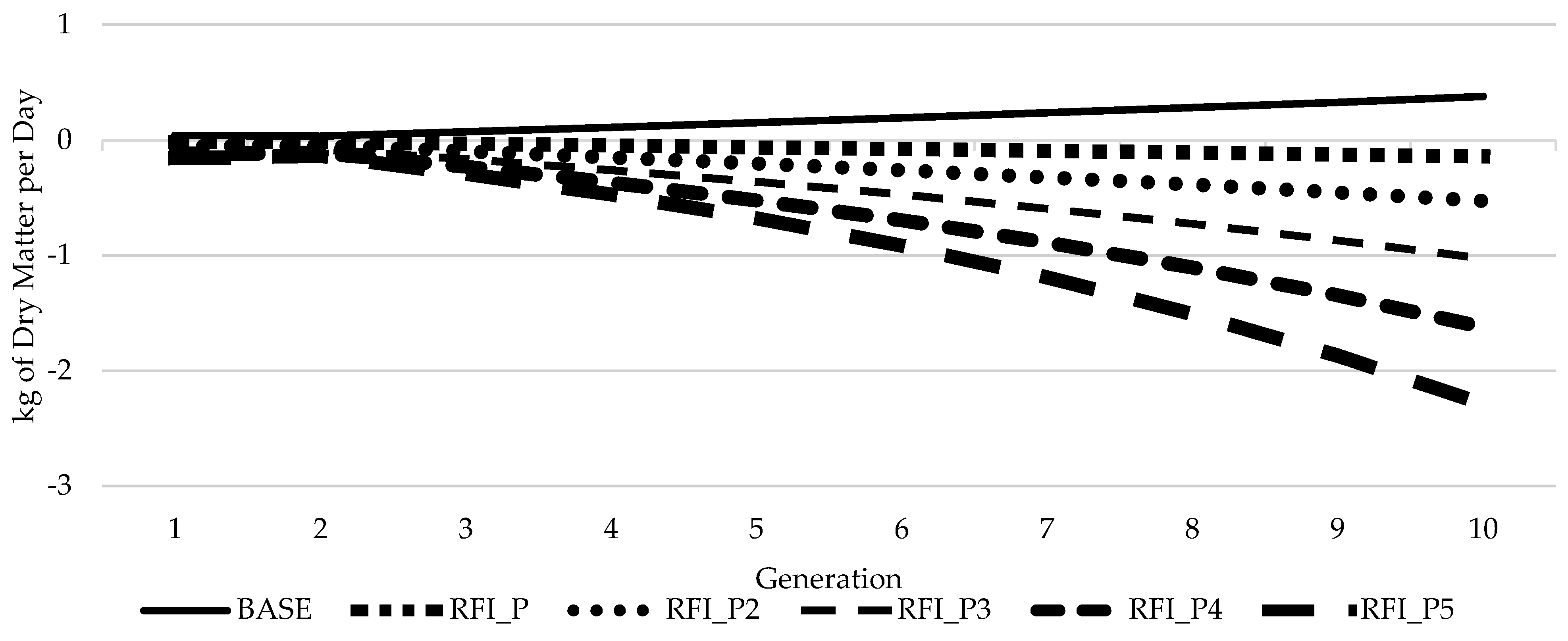
3.3. Response of Feed Efficiency over Time
4. Discussion
4.1. Genetic Parameters
4.2. Response to Selection of Feed Efficiency
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agriculture and Agri-Foods Canada. Advancing an Innovative and Competitive Dairy Sector; Agriculture and Agri-Foods Canada: Ottawa, ON, Canada, 2017.
- Connor, E.E. Invited review: Improving feed efficiency in dairy production: Challenges and possibilities. Animal 2015, 9, 395–408. [Google Scholar] [CrossRef] [Green Version]
- Van Biert, P. Economic of Milk Production in Alberta; Alberta Government: Edmonton, AB, Canada, 2018.
- Hemme, T.; Uddin, M.M.; Ndambi, O.A. Benchmarking cost of milk production in 46 countries. J. Rev. Glob. Econ. 2014, 3, 254–270. [Google Scholar] [CrossRef] [Green Version]
- Tempelman, R.J.; Spurlock, D.M.; Coffey, M.P.; Veerkamp, R.F.; Armentano, L.E.; Weigel, K.A.; de Haas, Y.; Staples, C.R.; Connor, E.E.; Lu, Y.; et al. Heterogeneity in genetic and nongenetic variation and energy sink relationships for residual feed intake across research stations and countries. J. Dairy Sci. 2015, 98, 2013–2026. [Google Scholar] [CrossRef] [Green Version]
- Pryce, J.E.; Gonzalez-Recio, O.; Nieuwhof, G.; Wales, W.J.; Coffey, M.P.; Hayes, B.J.; Goddard, M.E. Hot topic: Definition and implementation of a breeding value for feed efficiency in dairy cows. J. Dairy Sci. 2015, 98, 7340–7350. [Google Scholar] [CrossRef] [Green Version]
- Miglior, F.M.; Muir, B.L.; Van Doormaal, B.J. Selection indices in Holstein cattle of various countries. J. Dairy Sci. 2005, 88, 1255–1263. [Google Scholar] [CrossRef]
- Miglior, F.M.; Fleming, A.; Malchiodi, F.; Brito, L.F.; Martin, P.; Baes, C.F. A 100-Year Review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017, 100, 10251–10271. [Google Scholar] [CrossRef] [PubMed]
- Canadian Dairy Network. Canada Introduces Its New Profit Index: Pro$. Available online: https://www.cdn.ca/document.php?id=393 (accessed on 19 April 2019).
- Canadian Dairy Commission Production. Available online: http://www.cdc-ccl.gc.ca/CDC/index-eng.php?id=3801 (accessed on 22 May 2019).
- Kennedy, B.W.; Van Der Werf, J.H.J.; Meuwissent, T.H.E. Genetic and Statistical Properties of Residual Feed Intake. J. Anim. Sci. 1993, 71, 3239–3250. [Google Scholar] [CrossRef] [PubMed]
- Patience, J.F.; Rossoni-Serão, M.C.; Gutiérrez, N.A. A review of feed efficiency in swine: Biology and application. J. Anim. Sci. Biotechnol. 2015, 6, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdalla, E.E.A.; Schenkel, F.S.; Begli, H.E.; Willems, O.; van As, P.; Vanderhout, R.; Wood, B.J.; Baes, C.F. Single-Step Methodology for Genomic Evaluation in Turkeys (Meleagris gallopavo). Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Liinamo, A.E.; Mäntysaari, P.; Mäntysaari, E.A. Short communication: Genetic parameters for feed intake, production, and extent of negative energy balance in Nordic Red dairy cattle. J. Dairy Sci. 2012, 95, 6788–6794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berry, D.P.; Coffey, M.P.; Pryce, J.E.; de Haas, Y.; Løvendahl, P.; Krattenmacher, N.; Crowley, J.J.; Wang, Z.; Spurlock, D.M.; Weigel, K.A.; et al. International genetic evaluations for feed intake in dairy cattle through the collation of data from multiple sources. J. Dairy Sci. 2014, 97, 3894–3905. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Recio, O.; Pryce, J.E.; Haile-Mariam, M.; Hayes, B.J. Incorporating heifer feed efficiency in the Australian selection index using genomic selection. J. Dairy Sci. 2014, 97, 3883–3893. [Google Scholar] [CrossRef]
- Veerkamp, R.F.; Calus, M.P.L.; de Jong, G.; van der Linde, R.; de Haas, Y. Breeding values for dry matter intake for Dutch bulls based on DGV for DMI and BV for predictors. In Proceedings of the Page in the 10th World Congress on Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014. [Google Scholar]
- Løvendahl, P.; Difford, G.F.; Li, B.; Chagunda, M.G.G.; Huhtanen, P.; Lidauer, M.H.; Lassen, J.; Lund, P. Review: Selecting for improved feed efficiency and reduced methane emissions in dairy cattle. Animal 2018, 12 (Suppl. S2), s336–s349. [Google Scholar] [CrossRef] [Green Version]
- McParland, S.; Lewis, E.; Kennedy, E.; Moore, S.G.; McCarthy, B.; O’Donovan, M.; Butler, S.T.; Pryce, J.E.; Berry, D.P. Mid-infrared spectrometry of milk as a predictor of energy intake and efficiency in lactating dairy cows. J. Dairy Sci. 2014, 97, 5863–5871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassen, J.; Thomasen, J.; Hansen, R.H.; Nielsen, G.; Olsen, E.; Stentebjerg, P.R.B.; Hansen, N.; Borchersen, S. Individual measure of feed intake on in-house commercial dairy cattle using 3D camera system. In Proceedings of the 11th World Congress on Genetics Applied to Livestock Production, Volume Technologies–Novel Phenotypes, Auckland, New Zealand, 11–16 February 2018. [Google Scholar]
- Seymour, D.J.; Cánovas, A.; Baes, C.F.; Chud, T.C.S.; Osborne, V.R.; Cant, J.P.; Brito, L.F.; Gredler-Grandl, B.; Finocchiaro, R.; Veerkamp, R.F.; et al. Invited review: Determination of large-scale individual dry matter intake phenotypes in dairy cattle. J. Dairy Sci. 2019, 102, 7655–7663. [Google Scholar] [CrossRef] [PubMed]
- Brito, L.F.; Rojas de Oliveira, H.; Houlahan, K.; Fonseca, P.A.S.; Lam, S.; Butty, A.M.; Seymour, D.J.; Vargas, G.A.; Chud, T.C.S.; Silva, F.F.; et al. Invited Review: Genetic mechanisms underlying feed utilization and implementation of genomic selection for improved feed efficiency in dairy cattle. Can. J. Anim. Sci. 2020, 100, 587–604. [Google Scholar] [CrossRef]
- Berry, D.P.; Crowley, J.J. Cell Biology Symposium: Genetics of feed efficiency in dairy and beef cattle 1. J. Anim. Sci 2013, 91, 1594–1613. [Google Scholar] [CrossRef] [Green Version]
- Sjaunja, L.O.; Bævre, L.; Junkkarinen, L.; Pedersen, J.; Setälä, J. A nordic proposal for an energy corrected milk (ECM) formula. In Proceedings of the International Committee for Recording the Productivity of Milk Aniamals, Paris, France, 2–6 July 1990. [Google Scholar]
- Oliveira, G.A.; Schenkel, F.S.; Alcantara, L.; Houlahan, K.; Lynch, C.; Baes, C.F. Estimated genetic parameters for currently genetically evaluated traits in Canadian Holsteins. J. Dairy Sci. 2021, in press. [Google Scholar]
- Miglior, F.M.; Baes, C.F.; Cánovas, A.; Coffey, M.P.; Connor, E.E.; De Pauw, M.; Goddard, E.; Hailu, G.; Lassen, J.; Malchiodi, F.; et al. A progress report for the Efficient Dairy Genome Project. In Proceedings of the 2018 ASAS-CSAS Annual Meeting, Vancouver, BC, Canada, 8–12 July 2018. [Google Scholar]
- Pryce, J.E.; Wales, W.J.; De Haas, Y.; Veerkamp, R.F.; Hayes, B.J. Genomic selection for feed efficiency in dairy cattle. Animal 2014, 8, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K. WOMBAT—A tool for mixed model analyses in quantitative genetics by restricted maximum likelihood (REML). J. Zhejiang Univ. Sci. B 2007, 8, 815–821. [Google Scholar] [CrossRef] [Green Version]
- Täubert, H.; Reinhardt, F.; Simianer, H. ZPLAN+ A new software to evaluate and optimize animal breeding programs. In Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, 1–6 August 2010; pp. 1–6. [Google Scholar]
- Hazel, L.N.; Lush, J.L. The efficiency of three methods of selection. J. Hered. 1942, 33, 393–399. [Google Scholar] [CrossRef]
- Hill, W.G. Prediction and evaluation of response to selection with overlapping generations. Anim. Prod. 1974, 18, 117–139. [Google Scholar] [CrossRef]
- Van Doormaal, B.J. Impact of Genomics on Genetic Selection and Gain; Canadian Dairy Network: Guelph, ON, Canada, 2014. [Google Scholar]
- Schaeffer, L.R. Strategy for applying genome wide selection in dairy cattle. J. Anim. Breed. Genet. 2006, 123, 218–223. [Google Scholar] [CrossRef]
- Van Doormaal, B.J. Genomic Young Bulls: Accelerating Genetic Progress. Available online: https://www.cdn.ca/images/uploaded/file/GenomicYoungBullsArticle-September2017.pdf (accessed on 21 January 2019).
- Wilton, J.W.; Quinton, V.; Quinton, C. Optimizing Animal Genetic Improvement, 1st ed.; Centre for the Genetic Improvement of Livestock: Guelph, ON, Canada, 2013. [Google Scholar]
- Van der Werf, J.H.J. Teaching Software for Quantitative Genetics at UNE. Available online: https://jvanderw.une.edu.au/software.htm (accessed on 22 January 2019).
- Lang, B. The Cost of Raising Replacement Dairy Heifers. Available online: http://www.omafra.gov.on.ca/english/livestock/dairy/facts/11-055.htm (accessed on 24 April 2018).
- Cabrera, V.E. Economics of fertility in high-yielding dairy cows on confined TMR systems. Animal 2014, 8, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gohary, K.; Overton, M.W.; Von Massow, M.; Leblanc, S.J.; Lissemore, K.D.; Duffield, T.F. The cost of a case of subclinical ketosis in Canadian dairy herds. Can. Vet. J. 2016, 5757, 728–732. [Google Scholar]
- McArt, J.; Nydam, D.V.; Overton, M.W. Hyperketonemia in early lactation dairy cattle: A deterministic estimate of component and total cost per case. J. Dairy Sci. 2015, 98, 2043–2054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, C.M.; Baes, C.F.; Amer, P.R.; Quinton, C.; Martin, P.; Osborne, V.R.; Pryce, J.E.; Miglior, F.M. Determining the economic value of daily dry matter intake and associated methane emissions in dairy cattle. Animal 2019, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dairy Farmers of Ontario. Producer Milk Prices. Available online: http://www.milk.org/Corporate/Producers/PrdMilkPrices.aspx (accessed on 20 August 2020).
- De Haas, Y.; Pryce, J.E.; Calus, M.P.L.; Wall, E.; Berry, D.P.; Løvendahl, P.; Krattenmacher, N.; Miglior, F.M.; Weigel, K.A.; Spurlock, D.M.; et al. Genomic prediction of dry matter intake in dairy cattle from an international data set consisting of research herds in Europe, North America, and Australasia. J. Dairy Sci. 2015, 98, 6522–6534. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Fikse, W.F.; Lassen, J.; Lidauer, M.H.; Løvendahl, P.; Mäntysaari, P.; Berglund, B. Genetic parameters for dry matter intake in primiparous Holstein, Nordic Red, and Jersey cows in the first half of lactation. J. Dairy Sci. 2016, 99, 7232–7239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Byskov, M.V.; Fogh, A.; Løvendahl, P. Genetic parameters of rumination time and feed efficiency traits in primiparous Holstein cows under research and commercial conditions. J. Dairy Sci. 2017, 100, 9635–9642. [Google Scholar] [CrossRef]
- Hardie, L.C.; VandeHaar, M.J.; Tempelman, R.J.; Weigel, K.A.; Armentano, L.E.; Wiggans, G.R.; Veerkamp, R.F.; de Haas, Y.; Coffey, M.P.; Connor, E.E.; et al. The genetic and biological basis of feed efficiency in mid-lactation Holstein dairy cows. J. Dairy Sci. 2017, 100, 9061–9075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negussie, E.; Mehtiö, T.; Mäntysaari, P.; Løvendahl, P.; Mäntysaari, E.A.; Lidauer, M.H. Reliability of breeding values for feed intake and feed efficiency traits in dairy cattle: When dry matter intake recordings are sparse under different scenarios. J. Dairy Sci. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzanilla Pech, C.I.V.; Veerkamp, R.F.; Calus, M.P.L.; Zom, R.; van Knegsel, A.; Pryce, J.E.; De Haas, Y. Genetic parameters across lactation for feed intake, fat- and protein-corrected milk, and liveweight in first-parity Holstein cattle. J. Dairy Sci. 2014, 97, 5851–5862. [Google Scholar] [CrossRef] [PubMed]
- Bilal, G.; Cue, R.I.; Hayes, J.F. Genetic and phenotypic associations of type traits and body condition score with dry matter intake, milk yield, and number of breedings in first lactation Canadian Holstein cows. Can. J. Anim. Sci. 2016, 96, 434–447. [Google Scholar] [CrossRef] [Green Version]
- Lin, Z.; Macleod, I.; Pryce, J.E. Short communication: Estimation of genetic parameters for residual feed intake and feeding behavior traits in dairy heifers. J. Dairy Sci. 2013, 96, 2654–2656. [Google Scholar] [CrossRef] [PubMed]
- Manafiazar, G.H.; Goonewardene, L.; Miglior, F.M.; Crews, D.H.; Basarab, J.A.; Okine, E.; Wang, Z. Genetic and phenotypic correlations among feed efficiency, production and selected conformation traits in dairy cows. Animal 2016, 10, 381–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Vandehaar, M.J.; Spurlock, D.M.; Weigel, K.A.; Armentano, L.E.; Staples, C.R.; Connor, E.E.; Wang, Z.; Bello, N.M.; Tempelman, R.J. An alternative approach to modeling genetic merit of feed efficiency in dairy cattle. J. Dairy Sci. 2015, 98, 6535–6551. [Google Scholar] [CrossRef] [Green Version]
- Seymour, D.J.; Cánovas, A.; Chud, T.C.S.; Cant, J.P.; Osborne, V.R.; Baes, C.F.; Schenkel, F.S.; Miglior, F.M. The dynamic behavior of feed efficiency in primiparous dairy cattle. J. Dairy Sci. 2020, 103, 1528–1540. [Google Scholar] [CrossRef]
- Rodney, R.M.; Celi, P.; Scott, W.; Breinhild, K.; Santos, J.E.P.; Lean, I.J. Effects of nutrition on the fertility of lactating dairy cattle. J. Dairy Sci. 2018, 101, 5115–5133. [Google Scholar] [CrossRef] [Green Version]
- Chud, T.C.S.; Miglior, F.; Seymour, D.J.; Hailemariam, D.; Baes, C.F.; Schenkel, F.S. Genetic parameters for dry matter intake, metabolic body weight, and energy corrected milk in dairy cattle. In Proceedings of the Abstracts of the 2019 American Dairy Science Association Annual Meeting, Cincinnati, OH, USA, 23–26 June 2019. [Google Scholar]
- Garnsworthy, P.C. Body Condition Score in Dairy Cows: Targets for Production and Fertility. Recent Adv. Anim. Nutr. 2011, 2006, 61–86. [Google Scholar] [CrossRef]
- Goff, J.P. The monitoring, prevention, and treatment of milk fever and subclinical hypocalcemia in dairy cows. Vet. J. 2008, 176, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Kupczyński, R.; Szumny, A.; Wujcikowska, K.; Pachura, N. Metabolism, ketosis treatment and milk production after using glycerol in dairy cows: A review. Animals 2020, 10, 1379. [Google Scholar] [CrossRef] [PubMed]
- Pryce, J.E.; Nguyen, T.T.T.; Axford, M.; Nieuwhof, G.; Shaffer, M. Symposium review: Building a better cow—The Australian experience and future perspectives. J. Dairy Sci. 2018, 101, 3702–3713. [Google Scholar] [CrossRef] [PubMed]
- Council on Dairy Cattle Breeding Feed Saved (FSAV). Trait Reference Sheet. Available online: https://www.uscdcb.com/wp-content/uploads/2020/11/CDCB-Reference-Sheet-Feed-Saved-12_2020.pdf (accessed on 14 February 2021).
| Trait | Definition | Genetic Standard Deviation | Phenotypic Standard Deviation | Heritability | GEBV Accuracy |
|---|---|---|---|---|---|
| FY | Fat yield (kg) during a 305 d lactation | 30.67 1 | 70.13 1 | 0.29 a,1 | 0.80 |
| PY | Protein yield (kg) during a 305 d lactation | 21.33 1 | 55.45 1 | 0.22 a,1 | 0.79 |
| BCS | The measure of the fat covering over the tail head and rump on a scale of 1 (very thin) to 5 (very fat) in first lactation | 0.15 1 | 0.35 1 | 0.23 a,1 | 0.77 |
| STAT | Measure (cm) from the top of the spine in between hips to ground in first lactation | 2.19 1 | 4.42 1 | 0.47 a,1 | 0.77 |
| AFS | Number of days from birth to first insemination | 10.91 1 | 56.08 1 | 0.04 a,1 | 0.69 |
| FSTC | Number of days from first service to conception in first lactation | 7.45 1 | 44.68 1 | 0.03 a,1 | 0.74 |
| CK | Binary scored trait (0:no case/unknown, 1:at least one case) in first lactation | 0.03 1 | 0.21 1 | 0.02 a,1 | 0.61 |
| DA | Binary scored trait (0:no case/unknown, 1:at least one case) in first lactation | 0.03 1 | 0.15 1 | 0.04 a,1 | 0.59 |
| DMI | Average dry matter intake per day for a 305 d lactation | 1.54 | 3.25 | 0.23 b | 0.59 2 |
| RFI | Average residual feed intake per day for a 305 d lactation | 0.89 | 2.50 | 0.13 b | 0.40 3 |
| -Sex | Bulls | Cows | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| -Time | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | |
| Bulls | 1 | 0 | 0.167 | 0.167 | 0.167 | 0 | 0 | 0.125 | 0.125 | 0.125 | 0.125 |
| 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Cows | 1 | 0 | 0.117 | 0.117 | 0.192 | 0.075 | 0 | 0.125 | 0.125 | 0.125 | 0.125 |
| 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | |
| 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | |
| 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| Trait- | - | FY | PY | BCS | STAT | AFS | FSTC | CK | DA |
|---|---|---|---|---|---|---|---|---|---|
| DMI | 0.43 ± 0.09 | 0.50 ± 0.09 | 0.14 ± 0.14 | 0.05 ± 0.13 | −0.61 ± 0.17 | −0.13 ± 0.23 | −0.07 ± 0.21 | −0.13 ± 0.16 | |
| 0.29 ± 0.02 | 0.29 ± 0.02 | 0.01 ± 0.02 | 0.25 ± 0.04 | −0.05 ± 0.04 | 0.04 ± 0.04 | 0.23 ± 0.14 | 0.15 ± 0.17 | ||
| RFI | −0.07 ± 0.14 | 0.08 ± 0.14 | 0.35 ± 0.17 | −0.16 ± 0.15 | −0.41 ± 0.24 | −0.04 ± 0.29 | −0.09 ± 0.26 | −0.19 ± 0.21 | |
| 0.03 ± 0.03 | 0.03 ± 0.02 | 0.03 ± 0.02 | 0.08 ± 0.04 | −0.05 ± 0.04 | 0.05 ± 0.04 | −0.32 ± 0.16 | −0.31 ± 0.21 |
| Scenario | FY (kg) | PY (kg) | BCS (Score) | STAT (cm) | AFS (Days) | FSTC (Days) | CK (Case) | DA (Case) | DMI (kg) | RFI (kg) | Total Index Response (CAD) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BASE | 0.53 | 0.44 | −0.09 | 0.00 | −0.11 | 0.18 | −0.16 | 0.05 | 0.26 | 0.04 | 206.44 |
| BASE_SD | 0.53 | 0.44 | −0.08 | 0.00 | −0.10 | 0.18 | −0.16 | 0.06 | 0.21 | 0.02 | 204.45 |
| DMI_C | 0.46 | 0.33 | −0.20 | 0.00 | 0.09 | 0.25 | −0.13 | 0.13 | 0.00 | −0.15 | 161.11 |
| DMI_P | 0.53 | 0.43 | −0.05 | 0.00 | −0.11 | 0.17 | −0.18 | 0.05 | 0.27 | 0.06 | 167.16 |
| DMI_N | 0.54 | 0.42 | −0.11 | 0.00 | −0.04 | 0.21 | −0.18 | 0.08 | 0.19 | −0.02 | 174.05 |
| DMI_C_SD | 0.44 | 0.28 | 0.05 | 0.00 | 0.02 | 0.11 | −0.29 | 0.04 | 0.00 | −0.09 | 167.12 |
| DMI_P_SD | 0.51 | 0.44 | −0.08 | 0.00 | −0.14 | 0.18 | −0.13 | 0.05 | 0.25 | 0.06 | 167.05 |
| DMI_N_SD | 0.53 | 0.41 | −0.11 | 0.00 | −0.05 | 0.19 | −0.18 | 0.07 | 0.13 | −0.03 | 183.04 |
| RFI_C | 0.53 | 0.41 | −0.05 | 0.01 | −0.07 | 0.17 | −0.21 | 0.06 | 0.22 | 0.00 | 196.57 |
| RFI_P | 0.54 | 0.44 | −0.16 | 0.02 | −0.07 | 0.20 | −0.13 | 0.08 | 0.21 | −0.02 | 211.58 |
| RFI_N | 0.51 | 0.43 | −0.02 | 0.00 | −0.14 | 0.16 | −0.18 | 0.03 | 0.30 | 0.10 | 194.81 |
| RFI_C_SD | 0.51 | 0.41 | 0.00 | 0.00 | −0.10 | 0.15 | −0.21 | 0.04 | 0.18 | 0.00 | 200.15 |
| RFI_P_SD | 0.53 | 0.44 | −0.15 | 0.00 | −0.08 | 0.19 | −0.13 | 0.07 | 0.17 | −0.02 | 211.19 |
| RFI_N_SD | 0.52 | 0.43 | −0.05 | 0.01 | −0.12 | 0.19 | −0.17 | 0.05 | 0.23 | 0.06 | 196.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Houlahan, K.; Schenkel, F.S.; Hailemariam, D.; Lassen, J.; Kargo, M.; Cole, J.B.; Connor, E.E.; Wegmann, S.; Junior, O.; Miglior, F.; et al. Effects of Incorporating Dry Matter Intake and Residual Feed Intake into a Selection Index for Dairy Cattle Using Deterministic Modeling. Animals 2021, 11, 1157. https://doi.org/10.3390/ani11041157
Houlahan K, Schenkel FS, Hailemariam D, Lassen J, Kargo M, Cole JB, Connor EE, Wegmann S, Junior O, Miglior F, et al. Effects of Incorporating Dry Matter Intake and Residual Feed Intake into a Selection Index for Dairy Cattle Using Deterministic Modeling. Animals. 2021; 11(4):1157. https://doi.org/10.3390/ani11041157
Chicago/Turabian StyleHoulahan, Kerry, Flavio S. Schenkel, Dagnachew Hailemariam, Jan Lassen, Morten Kargo, John B. Cole, Erin E. Connor, Silvia Wegmann, Oliveira Junior, Filippo Miglior, and et al. 2021. "Effects of Incorporating Dry Matter Intake and Residual Feed Intake into a Selection Index for Dairy Cattle Using Deterministic Modeling" Animals 11, no. 4: 1157. https://doi.org/10.3390/ani11041157
APA StyleHoulahan, K., Schenkel, F. S., Hailemariam, D., Lassen, J., Kargo, M., Cole, J. B., Connor, E. E., Wegmann, S., Junior, O., Miglior, F., Fleming, A., Chud, T. C. S., & Baes, C. F. (2021). Effects of Incorporating Dry Matter Intake and Residual Feed Intake into a Selection Index for Dairy Cattle Using Deterministic Modeling. Animals, 11(4), 1157. https://doi.org/10.3390/ani11041157







