Genetic Characterization of a Sheep Population in Oaxaca, Mexico: The Chocholteca Creole
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Sampling, and Obtainment of DNA
2.2. Genotyping through Microsatellites
2.3. Intra-Racial Genetic Diversity
2.4. Interracial Genetic Diversity
2.5. Genetic Structure
3. Results
3.1. Intra-Species Genetic Diversity
3.2. Interracial Genetic Diversity
3.3. Genetic Structure of the Oaxacan Creole Sheep
4. Discussion
4.1. Intra-Racial Genetic Diversity
4.2. Inter-Racial Genetic Diversity
4.3. Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Secretaría de Agricultura, Ganadería, Desarrollo Rural, Pesca y Alimentación (SAGARPA). 2016. Padrón Ganadero Nacional. Available online: https://inehrm.gob.mx/recursos/Libros/SAGARPA.pdf (accessed on 1 August 2020).
- Servicio de Información Agroalimentaria y Pesquera (SIAP). Acciones Y Programas Para La Producción Pecuaria. 2018. Available online: https://www.gob.mx/siap/acciones-y-programas/produccion-pecuaria (accessed on 1 August 2020).
- Hernández-Bautista, J.; Salinas-Rios, T.; Rodríguez-Magadán, H.M.; Aquino-Cleto, M.; Mariscal-Méndez, A.; Ortiz-Muñoz, I.Y. Características que determinan el sistema de producción ovina en el estado de Oaxaca, México. Rev. Mex. De Agroecosistemas 2017, 4, 38–47. [Google Scholar]
- Martínez-Peña, M.; Villagómez-Cortés, J.A.; Mora-Brito, A.H. Rentabilidad del sistema de producción ovina en el bajo Mixe, Oaxaca, México. Agrociencia 2018, 52, 107–122. [Google Scholar]
- Food and Agriculture Organization of the United Nations. The Stage of the World´s Biodiversity for Food and Agriculture; Bélanger, J., Pilling, D., Eds.; FAO Commission on genetics resources for Food and Agriculture Assessment: Rome, Italy, 2019; p. 572. [Google Scholar]
- Martínez, R.D. Prejuicios que afectan a bovinos y ovinos criollos en Argentina. Revista AICA 2015, 5, 26–35. [Google Scholar]
- Martínez, S.R.; Vásquez, R.R. Evaluación de la conservación y comportamiento productivo del banco de germoplasma de la especie ovina en Colombia. Anim. Genet. Resour. Inf. 2005, 36, 33–45. [Google Scholar]
- Alonso, A.R.; Ulloa-Arvisu, R.; Gayosso-Vázquez, A. Mitochondrial DNA sequence analisys of the Mexican creole sheep (Ovis aries) reveals a narrow Iberian maternal origin. Mitochondrial DNA Part A 2017, 28, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Quiroz, J.; Martinez, M.A.; Zaragoza, L.; Perezgrovas, R.; Vega-Pla, J.L.; Delgado, J.V. Genetic characterization of the autochthonous sheep populations from Chiapas, México. Livest. Sci. 2008, 116, 156–161. [Google Scholar] [CrossRef]
- Delgado, J.V.; Martínez, M.A.; Acosta, A.; Álvarez, L.A.; Armstrong, E.; Camacho, E.; Cañón, J.; Cortés, O.; Dunner, S.; Landi, V.; et al. Genetic characterization of Latin-America creole cattle using microsatellite markers. Anim. Genet. 2011, 43, 2–10. [Google Scholar] [CrossRef]
- Ocampo, R.; Cardona, H.; Martínez, R. Genetic diversity of Colombian sheep by microsatellite markers. Chileanjar 2015, 76, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Aguirre-Riofrio, L.; Maza-Tandazo, T.; Quezada-Padilla, M.; Albito-Balcazar, O.; Flores-González, A.; Camacho-Enriquez, O.; Martínez-Martínez, A.; BioGoat Consortium; Delgado-Bermejo, J.V. Genetic characterization of the “Chusca Lojana” a creole goat reared in Ecuador, and its relationship with other Goat Breeds. Animals 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Vivas, N.A.; Landi, V.; Muñoz, F.J.; Bustamante, M.Y.; Álvarez, L.F. Genetic diversity of Colombian Creole sheep. J. MVZ Cordoba 2020, 25, e2185. [Google Scholar]
- Yatkin, S.; Özdil, F.; Özkan, E.Ü.; Genc, S.; Kaplan, S.; Kemal, E.G.; Arat, S.; Ihsan, M.S. Genetic Characterization of native donkey (Equus asinus) populations of Turkey using microsatellite markers. Animals 2020, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. BioTechniques 1991, 10, 506–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.D.E. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection; University of Dublin: Dublin, Ireland, 2001. [Google Scholar]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.C.; Boyle, T.J.B. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg. J. Bot. 1997, 130, 129–157. [Google Scholar]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. Genetix: 4.05 Logiciel Sous WindowsTM Pour la Genetique Des Populations; Université de Montpellier: Montpellier, France, 2003. [Google Scholar]
- Raymond, M.; Rousset, F. GENEPOP (Version 1.2): Population genetics software for exact test and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Guo, S.W.; Thompson, E.A. Performing the exact test of Hardy-Weinberg proportions for multiple alleles. Biometrics 1992, 48, 361–372. [Google Scholar] [CrossRef]
- Wright, S. Theory of gene frequencies. In Evolution and genetics of populations; University of Chicago: Chicago, IL, USA, 1969; pp. 291–293. [Google Scholar]
- Nei, M.; Tajima, F.; Tateno, Y. Accuracy of Estimated Phylogenetic Trees from Molecular Data. II. Gene Frequency Data. J. Mol. Evol. 1983, 19, 153–170. [Google Scholar] [CrossRef]
- Langella, O. Populations 1.2.28 CNRS UPR9034 1999. Available online: http//www.cnrs-gif.fr/pge/bioinfo/populations/index.php (accessed on 21 October 2019).
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–949. [Google Scholar]
- Arriaga-Jordán, C.M.; Pedraza-Fuentes, A.M.; Nava-Bernal, E.G.; Chávez-Mejía, M.C.; Castelán-Ortega, O.A. Livestock agrodiversity of Mazahua smallholder Campesino system in the highlands of central México. Hum. Ecol. 2005, 33, 821–845. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO). The State of the World´s Animal Genetic Resources for Food and Agriculture; Dafydd Pilling, D., Rischkowsky, B., Eds.; FAO: Rome, Italy, 2007; p. 39. [Google Scholar]
- Delgado, J.V.; Camacho, M.E. A Latinoamerican experience in the conservation of zoogenetic resources and traditional management system. Ital. J. Anim. Sci. 2007, 6, 120–121. [Google Scholar] [CrossRef]
- Sabir, J.; Mutwakil, M.; EL-Hanafy, A.; Al-Hejin, A.; Sadek, M.A.; Abou-Alsoud, M.; Qureshi, M.; Saini, K.; Ahmed, M. Appling molecular tools for improving livestock performance: From DNA markers to next generation sequencing technologies. J. Food Agric. Environ. 2014, 12, 351–363. [Google Scholar]
- Food and Agriculture Organization of the Unated Nations/International Society of Animal Genetics (FAO/ISAG). Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans; FAO: Rome, Italy, 2004; p. 24. [Google Scholar]
- Zhong, T.; Han, J.; Guo, J. Genetic diversity of Chinese indigenous sheep breeds inferred from microsatellite markers. Small Rumin. Res. 2010, 90, 88–94. [Google Scholar] [CrossRef]
- Peña, S.; Martínez, M.A.; Villegas, C.E.; Aulicino, M.; Género, E.R.; Giovambattista, G.; Martínez, R.D. Caracterización Genética de cuatro poblaciones de ovinos criollos de Argentina. J. Bag 2017, 28, 43–55. [Google Scholar]
- Ochipinti, G.; Núñez, L.; Cazal, C.; Samudio, A.; Castro, L.; Ramírez, L.; León, D.; Martínez, M.A.; Oka-Obara, A.; Landi, V.; et al. Diversidad genética en ovejas de los humedades de la región oriental de Paraguay. Actas Iberoam. Conserv. Anim. 2012, 2, 227–230. [Google Scholar]
- Ceccobelli, S.; Karsli, T.; Di Lorenzo, P.; Marozzi, G.; Landi, V.; Sarti, F.M.; Sabbioni, A.; Lasagna, E. Genetic diversity of Cornigliese sheep breed using STR markers. Small Rumin. Res. 2015, 123, 62–69. [Google Scholar] [CrossRef]
- Diez-Tascón, C.; Littlejohn, R.P.; Almeida, P.A.R.; Crawford, A.M. Genetic variation within the Merino sheep breed: Analysis of closely related populations using microsatellites. Anim. Genet. 2000, 31, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Baumung, R.; Simianer, H.; Hoffmann, I. Genetic diversity studies in farm animals—A survey. J. Anim. Breed Genet. 2004, 121, 361–373. [Google Scholar] [CrossRef]
- Wright, S. Evolutions, and the Genetics of Populations. Vol 4. Variability within and among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Rendo, F.; Iriondo, M.; Jugo, B.M.; Mazón, L.I.; Aguirre, A.; Vicario, A.; Estonba, A. Tracking diversity and differentiation in six sheep breeds from the North Iberian Peninsula through DNA variation. Small Rumin. Res. 2004, 52, 195–202. [Google Scholar] [CrossRef]
- Rodero, A.; Delgado, J.V.; Rodero, E. Primitive Andalusian livestock and their implications in the discovery of America. Arch. Zootec. 1992, 41, 383–400. [Google Scholar]
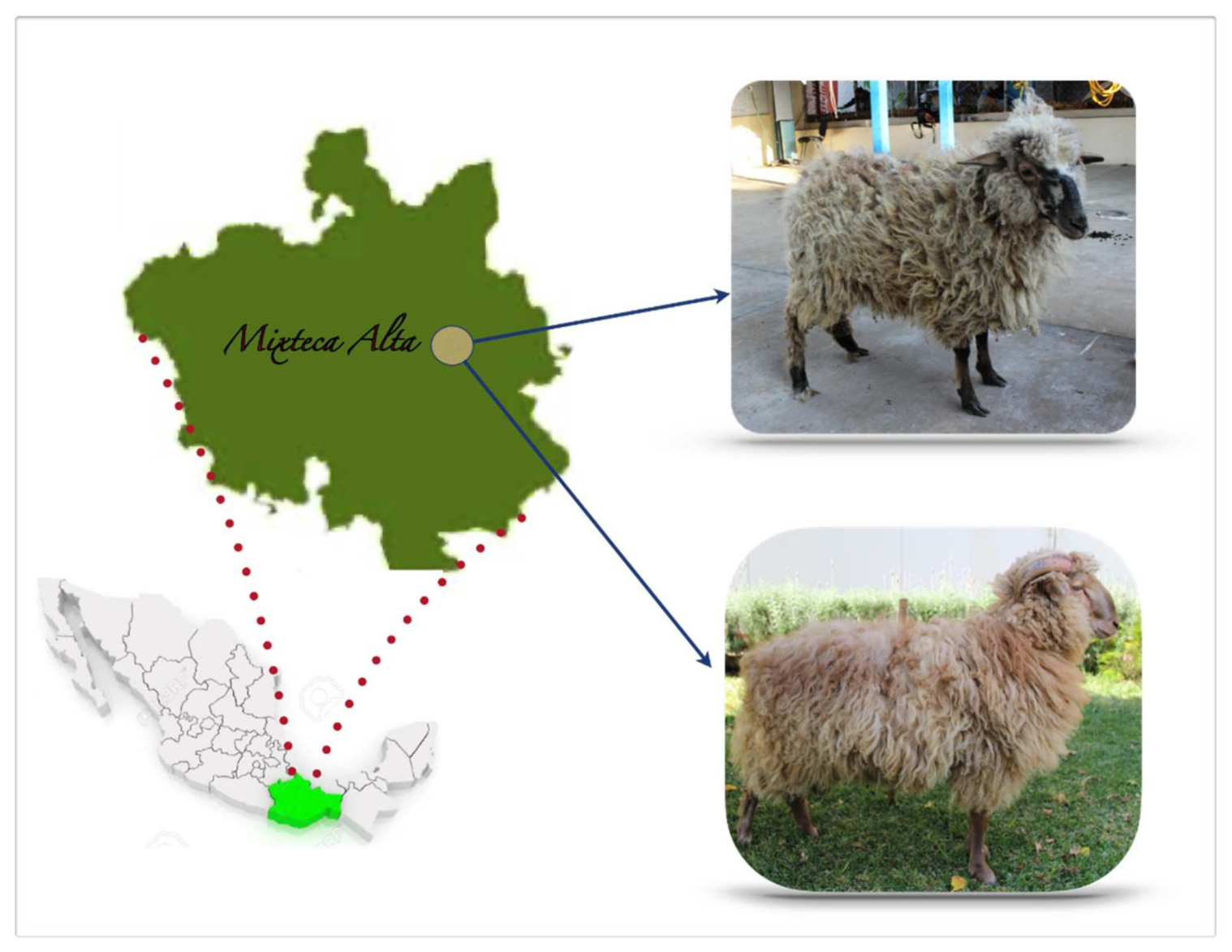
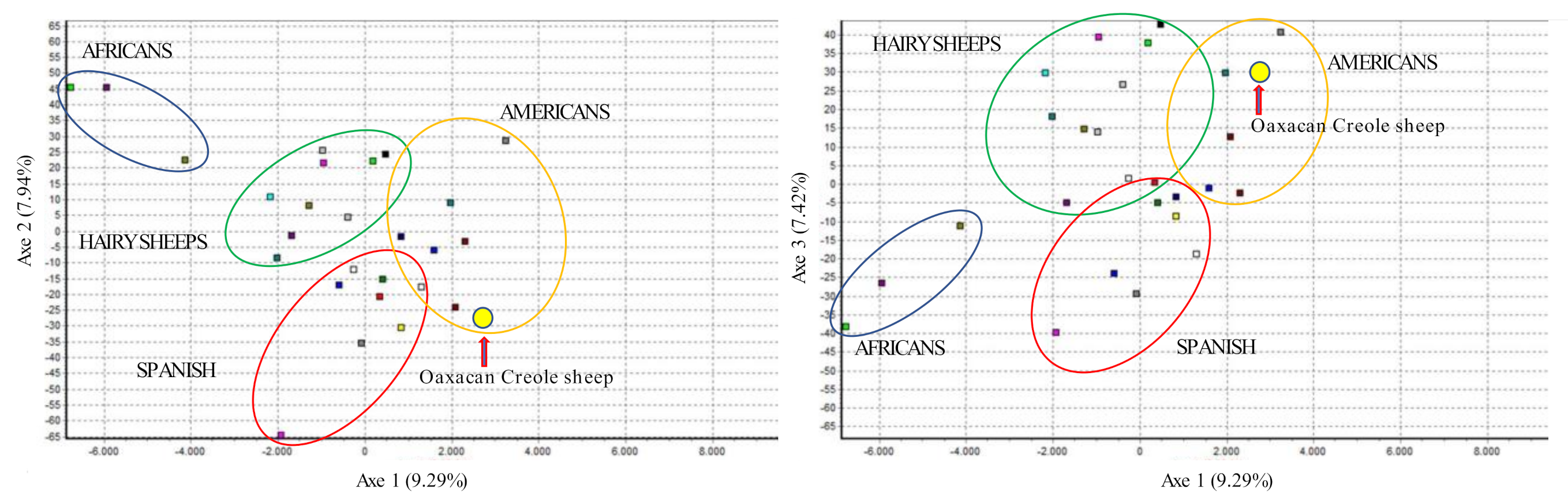
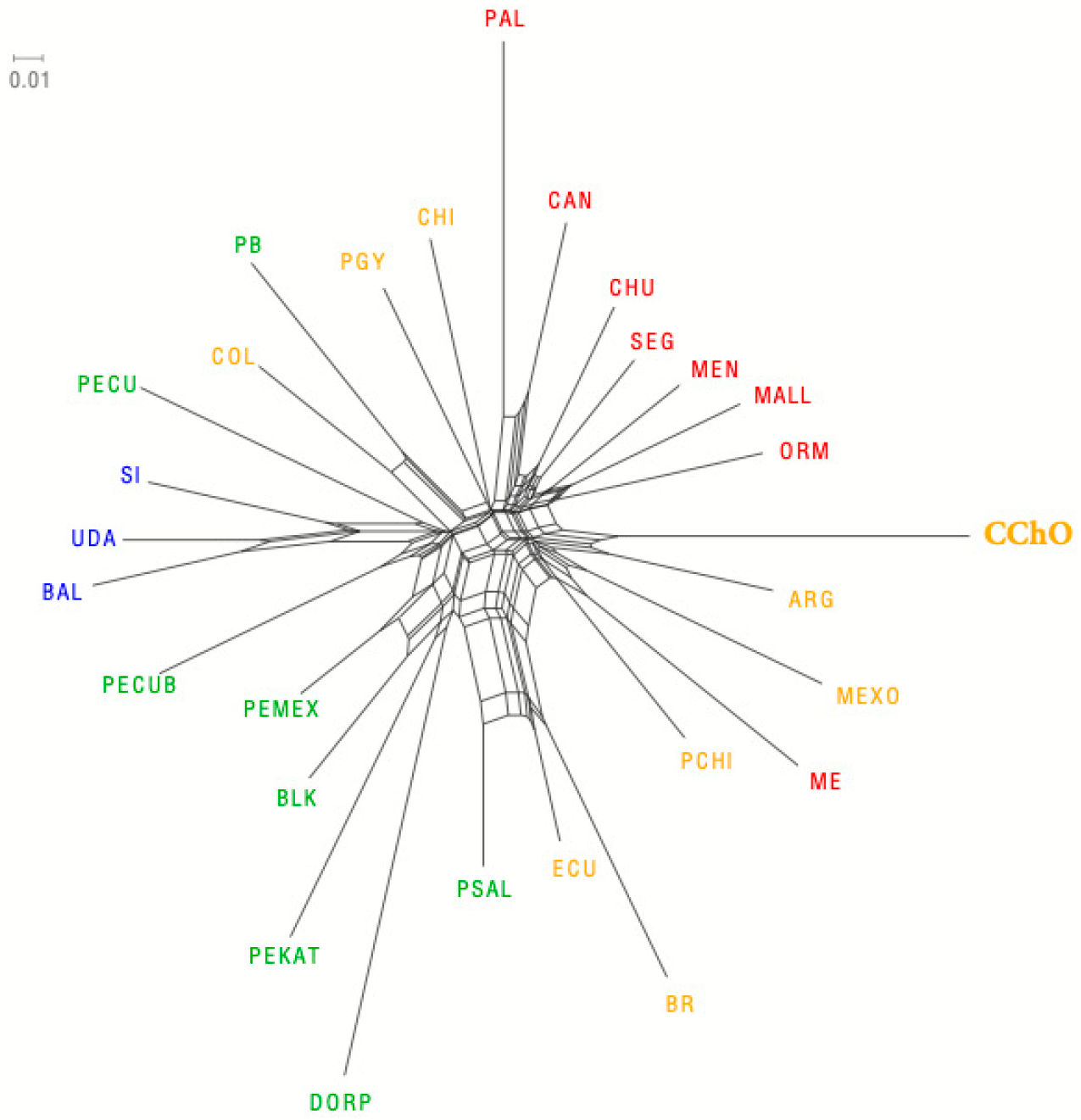
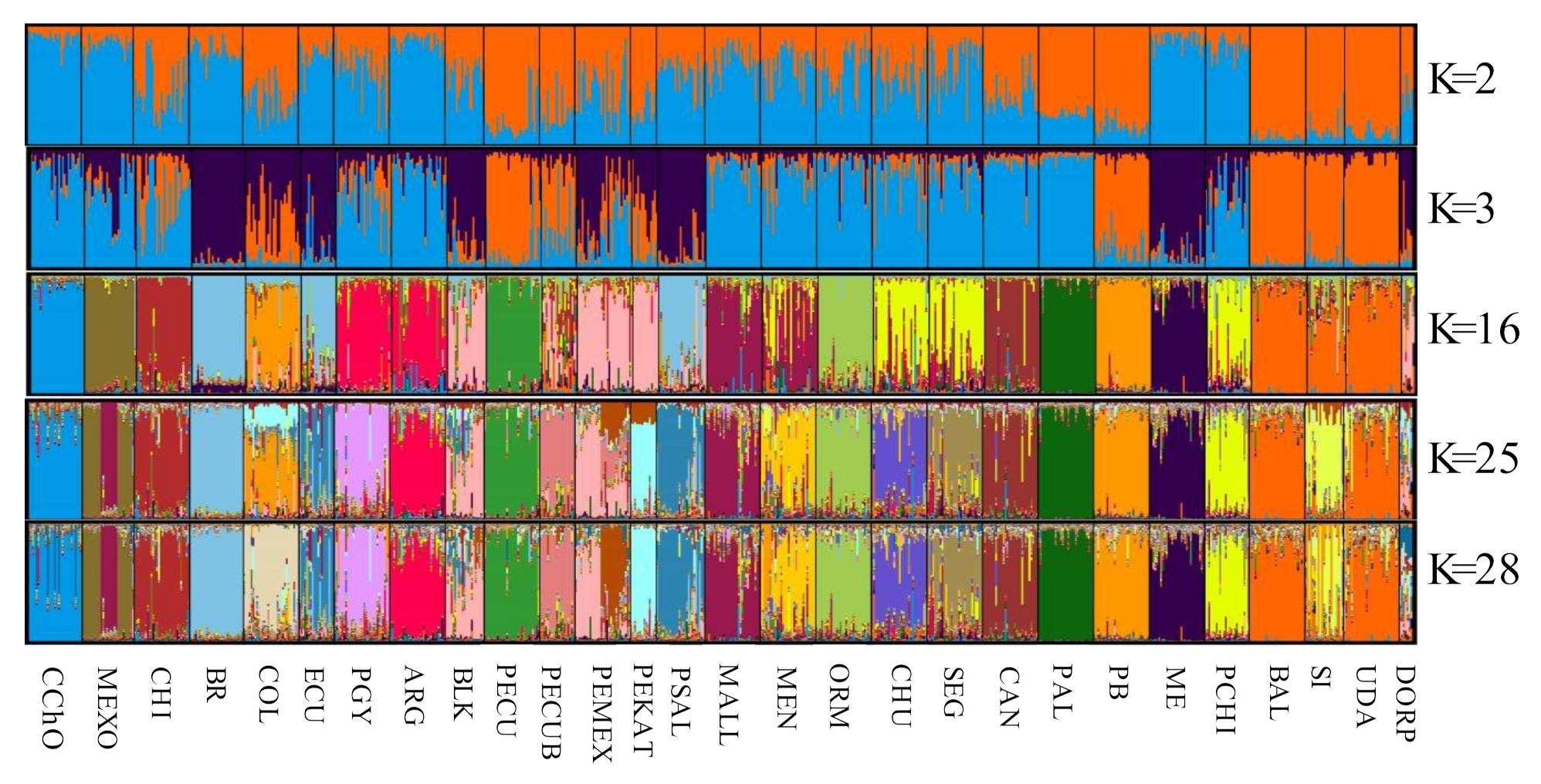
| Breed | Acronym | Provenance | n | |
|---|---|---|---|---|
| 1 | Creole Oaxaqueño | CChO | México | 29 |
| 2 | México | MEXO | México | 28 |
| 3 | Borrego de Chiapas | CHI | México | 30 |
| 4 | Criollo de Brasil | BR | Brasil | 29 |
| 5 | Criollo Colombiano | COL | Colombia | 30 |
| 6 | Criollo Ecuador | ECU | Ecuador | 19 |
| 7 | Criollo Paraguay | PGY | Paraguay | 30 |
| 8 | Criollo Argentina | ARG | Argentina | 30 |
| 9 | Blackbelly | BLK | México | 21 |
| 10 | Pelibuey Ecuador | PECU | Ecuador | 30 |
| 11 | Pelibuey Cubano | PECUB | Cuba | 19 |
| 12 | Pelibuey Mexicano | PEMEX | México | 30 |
| 13 | Katahdin | PEKAT | México | 14 |
| 14 | Pelibuey El Salvador | PSAL | El Salvador | 26 |
| 15 | Mallorquina | MALL | Spain | 30 |
| 16 | Menorquina | MEN | Spain | 30 |
| 17 | Roja Mallorquina | ORM | Spain | 30 |
| 18 | Churra | CHU | Spain | 30 |
| 19 | Segureño | SEG | Spain | 30 |
| 20 | Canaria | CAN | Spain | 30 |
| 21 | Palmera | PAL | Spain | 30 |
| 22 | Pelibuey | PB | Spain | 30 |
| 23 | Merino Español | ME | Spain | 30 |
| 24 | Merino Chileno | PCHI | Chile | 24 |
| 25 | Balami | BAL | Nigeria | 30 |
| 26 | Sidaun | SI | Sahara | 21 |
| 27 | UDA | UDA | Nigeria | 30 |
| 28 | Dorper | DORP | South Africa | 7 |
| Locus | Allele | Frequency | Locus | Allele | Frequency | Locus | Allele | Frequency |
|---|---|---|---|---|---|---|---|---|
| BM1818 | 1 | 7.14 | ILSTS008 | 1 | 57.14 | McM527 | 1 | 16.67 |
| 2 | 1.79 | 2 | 42.86 | 2 | 25 | |||
| 3 | 26.79 | ILSTS011 | 1 | 23.91 | 3 | 16.67 | ||
| 4 | 3.57 | 2 | 13.04 | 4 | 16.67 | |||
| 5 | 21.43 | 3 | 63.04 | 5 | 12.5 | |||
| 6 | 16.07 | ILSTS087 | 1 | 29.63 | 6 | 12.5 | ||
| 7 | 19.64 | 2 | 22.22 | OarAE129 | 1 | 8.93 | ||
| 8 | 3.57 | 3 | 12.96 | 2 | 48.21 | |||
| BM1824 | 1 | 26.09 | 4 | 11.11 | 3 | 32.14 | ||
| 2 | 19.57 | 5 | 24.07 | 4 | 10.71 | |||
| 3 | 4.35 | INRA005 | 1 | 35.42 | OarCP20 | 1 | 10.34 | |
| 4 | 50 | 2 | 29.17 | 2 | 32.76 | |||
| BM6506 | 1 | 32.76 | 3 | 8.33 | 3 | 56.9 | ||
| 2 | 34.48 | 4 | 14.58 | OarCP34 | 1 | 54.17 | ||
| 3 | 6.9 | 5 | 12.5 | 2 | 6.25 | |||
| 4 | 25.86 | INRA006 | 1 | 56.9 | 3 | 12.5 | ||
| BM6526 | 1 | 56.9 | 2 | 27.59 | 4 | 27.08 | ||
| 2 | 1.72 | 3 | 3.45 | OarCP49 | 1 | 5.56 | ||
| 3 | 17.24 | 4 | 12.07 | 2 | 12.96 | |||
| 4 | 20.69 | INRA023 | 1 | 58.93 | 3 | 29.63 | ||
| 5 | 3.45 | 2 | 17.86 | 4 | 18.52 | |||
| BM8125 | 1 | 20.69 | 3 | 16.07 | 5 | 11.11 | ||
| 2 | 18.97 | 4 | 5.36 | 6 | 1.85 | |||
| 3 | 8.62 | 5 | 1.79 | 7 | 5.56 | |||
| 4 | 51.72 | INRA035 | 1 | 6.9 | 8 | 12.96 | ||
| CD5 | 1 | 28 | 2 | 60.34 | 9 | 1.85 | ||
| 2 | 42 | 3 | 12.07 | OarFCB11 | 1 | 8.62 | ||
| 3 | 10 | 4 | 5.17 | 2 | 5.17 | |||
| 4 | 8 | 5 | 15.52 | 3 | 41.38 | |||
| 5 | 2 | INRA049 | 1 | 53.45 | 4 | 8.62 | ||
| 6 | 8 | 2 | 34.48 | 5 | 6.9 | |||
| 7 | 2 | 3 | 10.34 | 6 | 29.31 | |||
| CSRD247 | 1 | 3.45 | 4 | 1.72 | OarFCB20 | 1 | 6.82 | |
| 2 | 27.59 | INRA063 | 1 | 62.5 | 2 | 20.45 | ||
| 3 | 39.66 | 2 | 3.57 | 3 | 31.82 | |||
| 4 | 6.9 | 3 | 14.29 | 4 | 4.55 | |||
| 5 | 1.72 | 4 | 5.36 | 5 | 36.36 | |||
| 6 | 20.69 | 5 | 10.71 | OarFCB304 | 1 | 70.69 | ||
| CSSM66 | 1 | 1.72 | 6 | 3.57 | 2 | 3.45 | ||
| 2 | 6.9 | INRA132 | 1 | 8.33 | 3 | 25.86 | ||
| 3 | 24.14 | 2 | 20.83 | RM006 | 1 | 51.72 | ||
| 4 | 24.14 | 3 | 10.42 | 2 | 1.72 | |||
| 5 | 3.45 | 4 | 4.17 | 3 | 31.03 | |||
| 6 | 24.14 | 5 | 16.67 | 4 | 15.52 | |||
| 7 | 15.52 | 6 | 2.08 | SPS113 | 1 | 12.96 | ||
| D5S2 | 1 | 36.54 | 7 | 29.17 | 2 | 14.81 | ||
| 2 | 7.69 | 8 | 8.33 | 3 | 25.93 | |||
| 3 | 38.46 | INRA172 | 1 | 7.41 | 4 | 46.3 | ||
| 4 | 17.31 | 2 | 70.37 | SPS115 | 1 | 2.08 | ||
| ETH010 | 1 | 87.5 | 3 | 1.85 | 2 | 33.33 | ||
| 2 | 12.5 | 4 | 20.37 | 3 | 4.17 | |||
| ETH225 | 1 | 46.3 | MAF065 | 1 | 11.54 | 4 | 58.33 | |
| 2 | 48.15 | 2 | 7.69 | 5 | 2.08 | |||
| 3 | 3.7 | 3 | 34.62 | TGLA053 | 1 | 22.92 | ||
| 4 | 1.85 | 4 | 30.77 | 2 | 4.17 | |||
| HSC | 1 | 13.64 | 5 | 11.54 | 3 | 18.75 | ||
| 2 | 4.55 | MAF214 | 1 | 11.54 | 4 | 2.08 | ||
| 3 | 15.91 | 2 | 46.15 | 5 | 2.08 | |||
| 4 | 6.82 | 3 | 26.92 | 6 | 10.42 | |||
| 5 | 22.73 | 4 | 15.38 | 7 | 39.58 | |||
| 6 | 27.27 | McM042 | 1 | 4.35 | TGLA122 | 1 | 27.78 | |
| 7 | 9.09 | 2 | 39.13 | 2 | 12.96 | |||
| ILSTS005 | 1 | 12.96 | 3 | 30.43 | 3 | 33.33 | ||
| 2 | 31.48 | 4 | 4.35 | 4 | 20.37 | |||
| 3 | 16.67 | 5 | 21.74 | 5 | 5.56 | |||
| 4 | 11.11 | TGLA126 | 1 | 6.9 | ||||
| 5 | 27.78 | 2 | 27.59 | |||||
| 3 | 20.69 | |||||||
| 4 | 18.97 | |||||||
| 5 | 6.9 | |||||||
| 6 | 15.52 | |||||||
| 7 | 3.54 |
| Microsatellite | N° Allele | Ae | He | Ho | PIC | FIS | FIS IC | HWEd |
|---|---|---|---|---|---|---|---|---|
| ETH010 | 2 | 1.280 | 0.25 | 0.223 | 0.195 | −0.125 | (−0.244–−0.037) | 1 |
| OarFCB304 | 3 | 1.761 | 0.31 | 0.44 | 0.364 | 0.29805 | (−0.050–0.612) | 0.0315 * |
| SPS115 | 5 | 2.203 | 0.375 | 0.558 | 0.468 | 0.33226 | (−0.031–0.633) | 0.0256 * |
| ILSTS008 | 2 | 1.960 | 0.429 | 0.499 | 0.37 | 0.14286 | (−0.242–0.496) | 0.6978 |
| INRA172 | 4 | 1.843 | 0.444 | 0.466 | 0.41 | 0.04733 | (−0.181–0.225) | 0.0082 * |
| RM006 | 4 | 2.576 | 0.517 | 0.623 | 0.542 | 0.1716 | (−0.094–0.414) | 0.1545 |
| BM8125 | 4 | 2.827 | 0.517 | 0.658 | 0.596 | 0.21642 | (−0.085–0.479) | 0.0457 |
| ETH225 | 4 | 2.233 | 0.519 | 0.563 | 0.451 | 0.07965 | (−0.270–0.389) | 0.8494 |
| ILSTS011 | 3 | 2.120 | 0.522 | 0.54 | 0.467 | 0.03473 | (−0.282–0.348) | 0.7858 |
| INRA049 | 4 | 2.406 | 0.552 | 0.595 | 0.508 | 0.07342 | (−0.191–0.304) | 0.007 |
| INRA023 | 5 | 2.450 | 0.571 | 0.603 | 0.548 | 0.05263 | (−0.134–0.204) | 0.0466 * |
| INRA035 | 5 | 2.438 | 0.586 | 0.6 | 0.555 | 0.02359 | (−0.224–0.252) | 0.5145 |
| OarCP20 | 3 | 2.264 | 0.621 | 0.568 | 0.48 | −0.09446 | (−0.379–0.174) | 0.665 |
| CSRD247 | 6 | 3.541 | 0.621 | 0.73 | 0.67 | 0.15223 | (−0.081–0.357) | 0.0043 * |
| OarAE129 | 4 | 2.815 | 0.643 | 0.656 | 0.583 | 0.02115 | (−0.212–0.231) | 0.4807 |
| INRA006 | 4 | 2.406 | 0.655 | 0.595 | 0.523 | −0.10373 | (−0.310–0.062) | 0.0156 |
| D5S2 | 4 | 3.152 | 0.692 | 0.696 | 0.623 | 0.00552 | (−0.235–0.220) | 0.4133 |
| BM1824 | 4 | 2.792 | 0.696 | 0.656 | 0.582 | −0.06184 | (−0.383–0.226) | 0.1587 |
| SPS113 | 4 | 3.122 | 0.704 | 0.693 | 0.628 | −0.01646 | (−0.266–0.204) | 0.1321 |
| ILSTS87 | 5 | 4.459 | 0.704 | 0.79 | 0.739 | 0.11151 | (−0.120–0.316) | 0.6321 |
| OarCP34 | 4 | 2.589 | 0.708 | 0.627 | 0.556 | −0.13333 | (−0.431–0.148) | 0.3552 |
| INRA005 | 5 | 3.932 | 0.708 | 0.762 | 0.705 | 0.07126 | (−0.195–0.304) | 0.1144 |
| INRA063 | 6 | 2.337 | 0.714 | 0.582 | 0.542 | −0.23147 | (−0.366–−0.114) | 0.8619 |
| HSC | 7 | 5.408 | 0.727 | 0.834 | 0.79 | 0.13066 | (−0.108–0.327) | 0.0015 * |
| INRA132 | 8 | 5.460 | 0.75 | 0.834 | 0.793 | 0.10293 | (−0.098–0.277) | 0.0864 |
| OarFCB11 | 6 | 3.579 | 0.759 | 0.733 | 0.679 | −0.03529 | (−0.249–0.149) | 0.0817 |
| MAF065 | 6 | 4.024 | 0.769 | 0.766 | 0.713 | −0.00402 | (−0.192–0.158) | 0.4646 |
| ILSTS005 | 5 | 4.288 | 0.778 | 0.781 | 0.729 | 0.00456 | (−0.194–0.178) | 0.0044 * |
| McM042 | 5 | 3.369 | 0.783 | 0.719 | 0.649 | −0.09091 | (−0.318–0.110) | 0.7442 |
| BM6526 | 5 | 2.514 | 0.793 | 0.613 | 0.552 | −0.30101 | (−0.448–−0.174) | 0.4987 |
| MAF214 | 4 | 3.101 | 0.808 | 0.691 | 0.625 | −0.17318 | (−0.383–0.024) | 0.5427 |
| OarFCB20 | 5 | 3.546 | 0.818 | 0.735 | 0.668 | −0.11669 | (−0.327–0.072) | 0.6359 |
| BM1818 | 8 | 5.262 | 0.857 | 0.825 | 0.783 | −0.04013 | (−0.194–0.082) | 0.0793 |
| BM6506 | 4 | 3.357 | 0.862 | 0.714 | 0.644 | −0.21107 | (−0.417–−0.028) | 0.0482 * |
| CSSM66 | 7 | 4.875 | 0.862 | 0.809 | 0.764 | −0.06707 | (−0.228–0.075) | 0.6016 |
| CD5 | 7 | 3.592 | 0.88 | 0.736 | 0.682 | −0.2 | (−0.369–−0.048) | 0.9183 |
| OarCP49 | 9 | 5.718 | 0.889 | 0.841 | 0.804 | −0.05852 | (−0.192–0.045) | 0.0426 * |
| TGLA126 | 7 | 5.273 | 0.897 | 0.825 | 0.784 | −0.08901 | (−0.233–0.035) | 0.0247 * |
| McM527 | 6 | 5.647 | 0.917 | 0.84 | 0.798 | −0.09287 | (−0.234–0.037) | 0.5744 |
| TGLA053 | 7 | 3.879 | 0.958 | 0.758 | 0.704 | −0.27163 | (−0.419–−0.166) | 0.1518 |
| TGLA122 | 5 | 4.006 | 0.963 | 0.765 | 0.708 | −0.26592 | (−0.374–−0.180) | 0.1761 |
| Average | 5 | 3.327 | 0.685 | 0.671 | 0.609 | −0.022 | (−0.087–0.002) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salinas-Rios, T.; Hernández-Bautista, J.; Mariscal-Méndez, A.; Aquino-Cleto, M.; Martínez-Martínez, A.; Rodríguez-Magadán, H.M. Genetic Characterization of a Sheep Population in Oaxaca, Mexico: The Chocholteca Creole. Animals 2021, 11, 1172. https://doi.org/10.3390/ani11041172
Salinas-Rios T, Hernández-Bautista J, Mariscal-Méndez A, Aquino-Cleto M, Martínez-Martínez A, Rodríguez-Magadán HM. Genetic Characterization of a Sheep Population in Oaxaca, Mexico: The Chocholteca Creole. Animals. 2021; 11(4):1172. https://doi.org/10.3390/ani11041172
Chicago/Turabian StyleSalinas-Rios, Teodulo, Jorge Hernández-Bautista, Araceli Mariscal-Méndez, Magaly Aquino-Cleto, Amparo Martínez-Martínez, and Héctor Maximino Rodríguez-Magadán. 2021. "Genetic Characterization of a Sheep Population in Oaxaca, Mexico: The Chocholteca Creole" Animals 11, no. 4: 1172. https://doi.org/10.3390/ani11041172






