Blood Transcriptomics of Turbot Scophthalmus maximus: A Tool for Health Monitoring and Disease Studies
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Sampling
2.3. Histopathological, Immunochemical and PCR-Based Diagnosis
2.4. RNA-seq
3. Results
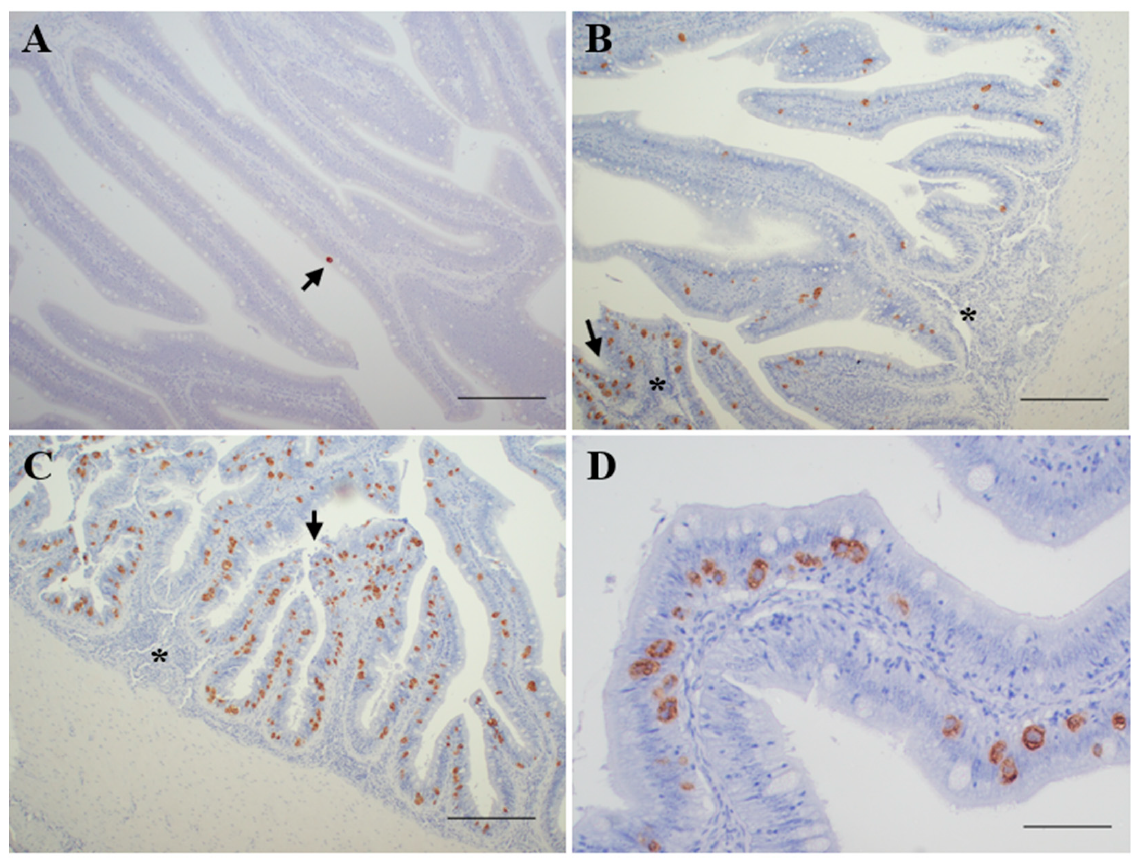
3.1. Diagnosis of Enteromyxosis
- Not infected (n = 8): negative detection by all the methods applied.
- Incipient infection (n = 14): only positive by PCR diagnosis.
- Slight infection (n = 15): positive by PCR and immunohistochemical diagnosis, showing reduced parasite burden and none or minimal histopathological lesions.
- Moderate infection (n = 13): positive by PCR, immunohistochemical and histological diagnosis, showing signs of enteritis and most of the intestinal folds parasitized, with variable parasite burden.
- Severe infection (n = 10): positive by PCR, immunohistochemical and histological diagnosis, showing high parasite burden and evident lesions of catarrhal enteritis.
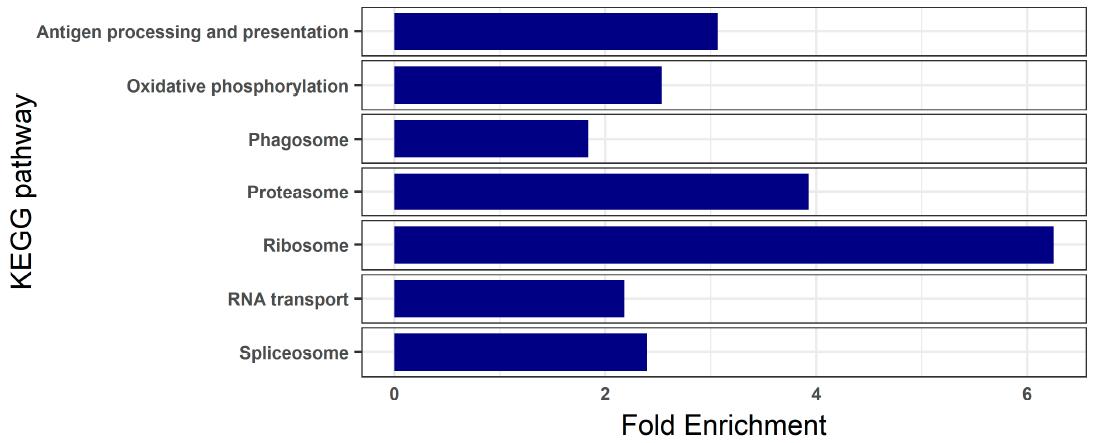
3.2. Blood Transcriptome of Healthy Turbot
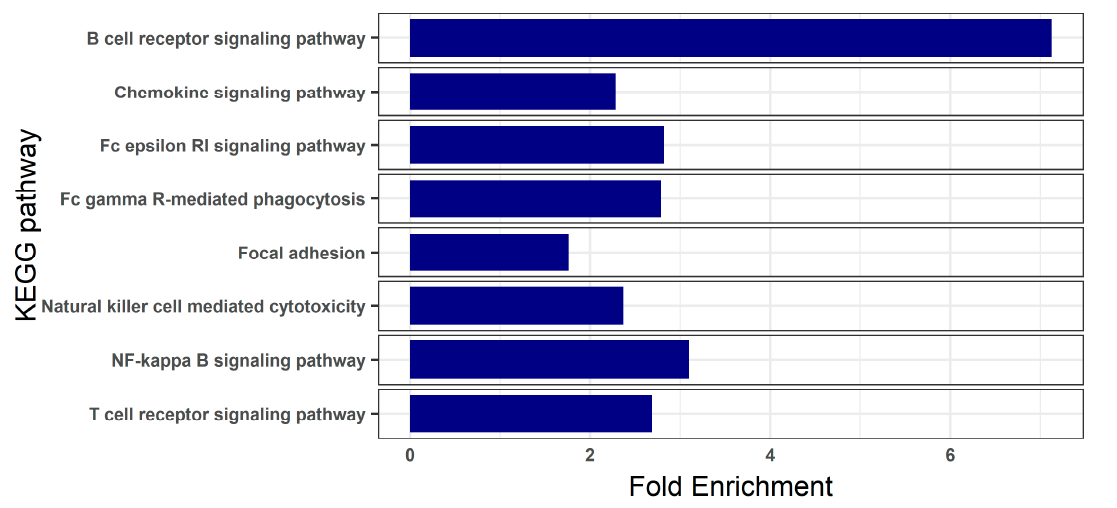
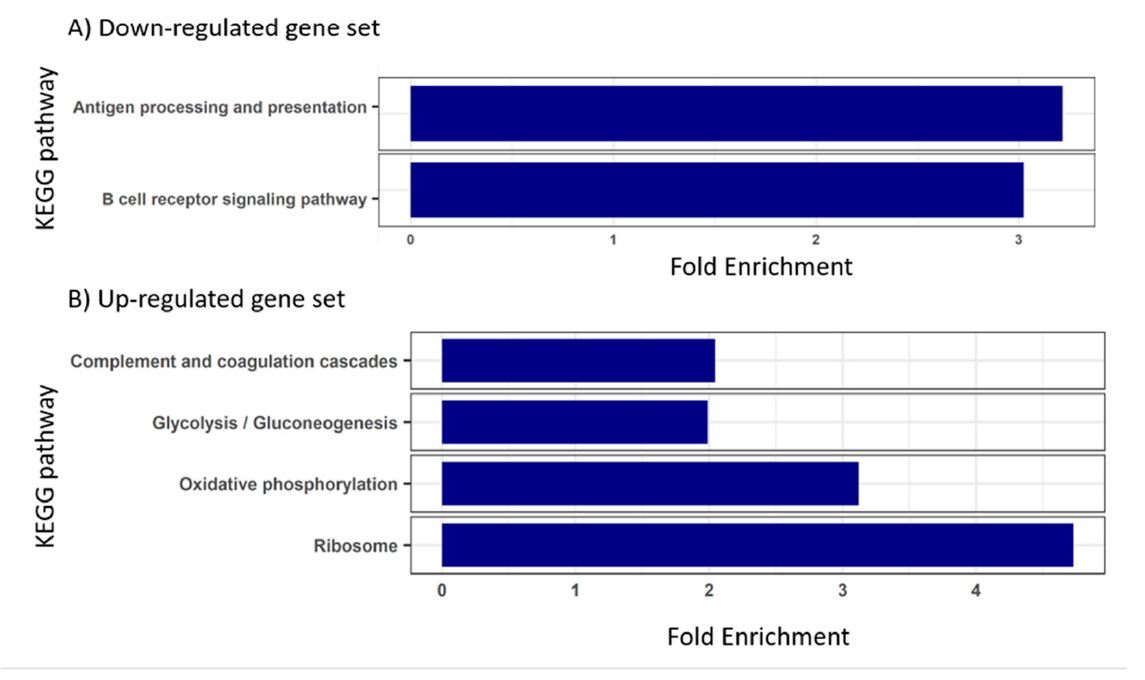
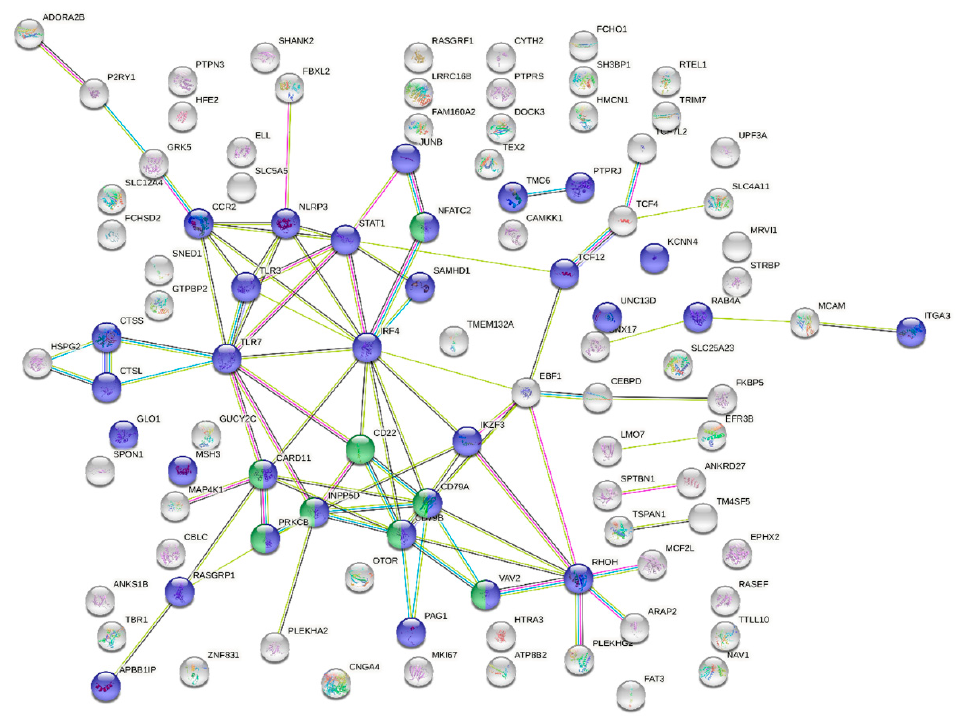
3.3. Blood Transcriptome during Enteromyxosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chaussabel, D.; Pascual, V.; Banchereau, J. Assessing the Human Immune System through Blood Transcriptomics. BMC Biol. 2010, 8, 84. [Google Scholar] [CrossRef] [PubMed]
- Mohr, S.; Liew, C.-C. The Peripheral-Blood Transcriptome: New Insights into Disease and Risk Assessment. Trends Mol. Med. 2007, 13, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Grant, K.R. Fish Hematology and Associated Disorders. Vet. Clin. N. Am. Exot. Anim. Pract. 2015, 18, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Aceves, M.A.; Lionetti, L.; Faggio, C. Multidisciplinary Haematology as Prognostic Device in Environmental and Xenobiotic Stress-Induced Response in Fish. Sci. Total Environ. 2019, 670, 1170–1183. [Google Scholar] [CrossRef] [PubMed]
- Chaussabel, D. Assessment of Immune Status Using Blood Transcriptomics and Potential Implications for Global Health. Semin. Immunol. 2015, 27, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Costa, V.; Aprile, M.; Esposito, R.; Ciccodicola, A. RNA-Seq and Human Complex Diseases: Recent Accomplishments and Future Perspectives. Eur. J. Hum. Genet. 2013, 21, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Cabantous, S.; Poudiougou, B.; Bergon, A.; Barry, A.; Oumar, A.A.; Traore, A.M.; Chevillard, C.; Doumbo, O.; Dessein, A.; Marquet, S. Understanding Human Cerebral Malaria through a Blood Transcriptomic Signature: Evidences for Erythrocyte Alteration, Immune/Inflammatory Dysregulation, and Brain Dysfunction. Available online: https://www.hindawi.com/journals/mi/2020/3280689/ (accessed on 27 October 2020).
- Blankley, S.; Berry, M.P.R.; Graham, C.M.; Bloom, C.I.; Lipman, M.; O’Garra, A. The Application of Transcriptional Blood Signatures to Enhance Our Understanding of the Host Response to Infection: The Example of Tuberculosis. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Sudhagar, A.; Kumar, G.; El-Matbouli, M. Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review. Int. J. Mol. Sci. 2018, 19, 245. [Google Scholar] [CrossRef]
- Li, Z.; Liu, X.; Liu, J.; Zhang, K.; Yu, H.; He, Y.; Wang, X.; Qi, J.; Wang, Z.; Zhang, Q. Transcriptome Profiling Based on Protein–Protein Interaction Networks Provides a Core Set of Genes for Understanding Blood Immune Response Mechanisms against Edwardsiella Tarda Infection in Japanese Flounder (Paralichthys olivaceus). Dev. Comp. Immunol. 2018, 78, 100–113. [Google Scholar] [CrossRef]
- Nombela, I.; Requena-Platek, R.; Morales-Lange, B.; Chico, V.; Puente-Marin, S.; Ciordia, S.; Mena, M.C.; Coll, J.; Perez, L.; Mercado, L.; et al. Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-Presenting Cell Markers. Cells 2019, 8, 386. [Google Scholar] [CrossRef]
- Jung, M.-H.; Chico, V.; Ciordia, S.; Mena, M.C.; Jung, S.-J.; Ortega-Villaizan, M.D.M. The Megalocytivirus RBIV Induces Apoptosis and MHC Class I Presentation in Rock Bream (Oplegnathus fasciatus) Red Blood Cells. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Dahle, M.K.; Wessel, Ø.; Timmerhaus, G.; Nyman, I.B.; Jørgensen, S.M.; Rimstad, E.; Krasnov, A. Transcriptome Analyses of Atlantic Salmon (Salmo salar L.) Erythrocytes Infected with Piscine Orthoreovirus (PRV). Fish Shellfish Immunol. 2015, 45, 780–790. [Google Scholar] [CrossRef]
- Pereiro, P.; Romero, A.; Diaz-Rosales, P.; Estepa, A.; Figueras, A.; Novoa, B. Nucleated Teleost Erythrocytes Play an Nk-Lysin- and Autophagy-Dependent Role in Antiviral Immunity. Front. Immunol. 2017, 8, 1458. [Google Scholar] [CrossRef]
- Ronza, P.; Robledo, D.; Bermúdez, R.; Losada, A.P.; Pardo, B.G.; Martínez, P.; Quiroga, M.I. Integrating Genomic and Morphological Approaches in Fish Pathology Research: The Case of Turbot (Scophthalmus maximus) Enteromyxosis. Front. Genet. 2019, 10, 26. [Google Scholar] [CrossRef]
- Ronza, P.; Robledo, D.; Losada, A.P.; Bermúdez, R.; Pardo, B.G.; Martínez, P.; Quiroga, M.I. The Teleost Thymus in Health and Disease: New Insights from Transcriptomic and Histopathological Analyses of Turbot, Scophthalmus maximus. Biology 2020, 9, 221. [Google Scholar] [CrossRef]
- Figueras, A.; Robledo, D.; Corvelo, A.; Hermida, M.; Pereiro, P.; Rubiolo, J.A.; Gomez-Garrido, J.; Carrete, L.; Bello, X.; Gut, M.; et al. Whole Genome Sequencing of Turbot (Scophthalmus maximus; Pleuronectiformes): A Fish Adapted to Demersal Life. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2016, 23, 181–192. [Google Scholar] [CrossRef]
- Redondo, M.J.; Palenzuela, O.; Álvarez-Pellitero, P. Studies on Transmission and Life Cycle of Enteromyxum scophthalmi (Myxozoa), an Enteric Parasite of Turbot Scophthalmus maximus. Folia Parasitol. 2004, 51, 188–198. [Google Scholar] [CrossRef]
- Bermúdez, R.; Losada, A.P.; Vázquez, S.; Redondo, M.J.; Álvarez-Pellitero, P.; Quiroga, M.I. Light and Electron Microscopic Studies on Turbot Psetta Maxima Infected with Enteromyxum scophthalmi: Histopathology of Turbot Enteromyxosis. Dis. Aquat. Org. 2010, 89, 209–221. [Google Scholar] [CrossRef]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS Server: A Web-Based Platform for Automated Annotation and Pathway Identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Quentel, C.; Obach, A. The Cellular Composition of the Blood and Haematopoietic Organs of Turbot Scophthalmus maximus L. J. Fish Biol. 1992, 41, 709–716. [Google Scholar] [CrossRef]
- Raghavachari, N.; Xu, X.; Munson, P.J.; Gladwin, M.T. Characterization of Whole Blood Gene Expression Profiles as a Sequel to Globin MRNA Reduction in Patients with Sickle Cell Disease. PLoS ONE 2009, 4, e6484. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shen, Y.; Wang, D.; Zhao, J.; Chen, X. Fish Red Blood Cells Express Immune Genes and Responses. Aquac. Fish. 2018, 3, 14–21. [Google Scholar] [CrossRef]
- Désert, C.; Merlot, E.; Zerjal, T.; Bed’hom, B.; Härtle, S.; Le Cam, A.; Roux, P.-F.; Baeza, E.; Gondret, F.; Duclos, M.J.; et al. Transcriptomes of Whole Blood and PBMC in Chickens. Comp. Biochem. Physiol. Part D Genom. Proteom. 2016, 20, 1–9. [Google Scholar] [CrossRef]
- Meitern, R.; Andreson, R.; Hõrak, P. Profile of Whole Blood Gene Expression Following Immune Stimulation in a Wild Passerine. BMC Genom. 2014, 15, 533. [Google Scholar] [CrossRef]
- Ganeshan, K.; Chawla, A. Metabolic Regulation of Immune Responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef]
- Ji, P.; Murata-Hori, M.; Lodish, H.F. Formation of Mammalian Erythrocytes: Chromatin Condensation and Enucleation. Trends Cell Biol. 2011, 21, 409–415. [Google Scholar] [CrossRef]
- Morera, D.; Roher, N.; Ribas, L.; Balasch, J.C.; Doñate, C.; Callol, A.; Boltaña, S.; Roberts, S.; Goetz, G.; Goetz, F.W.; et al. RNA-Seq Reveals an Integrated Immune Response in Nucleated Erythrocytes. PLoS ONE 2011, 6, e26998. [Google Scholar] [CrossRef]
- Houde, M.; Bertholet, S.; Gagnon, E.; Brunet, S.; Goyette, G.; Laplante, A.; Princiotta, M.F.; Thibault, P.; Sacks, D.; Desjardins, M. Phagosomes Are Competent Organelles for Antigen Cross-Presentation. Nature 2003, 425, 402–406. [Google Scholar] [CrossRef]
- Mantegazza, A.R.; Magalhaes, J.G.; Amigorena, S.; Marks, M.S. Presentation of Phagocytosed Antigens by MHC Class I and II. Traffic 2013, 14, 135–152. [Google Scholar] [CrossRef]
- Kloetzel, P.M. Antigen Processing by the Proteasome. Nat. Rev. Mol. Cell Biol. 2001, 2, 179–187. [Google Scholar] [CrossRef]
- Kloetzel, P.-M. The Proteasome and MHC Class I Antigen Processing. Biochim. Biophys. Acta Mol. Cell Res. 2004, 1695, 225–233. [Google Scholar] [CrossRef]
- Redondo, M.J.; Palenzuela, O.; Álvarez-Pellitero, P. In Vitro Studies on Viability and Proliferation of Enteromyxum scophthalmi (Myxozoa), an Enteric Parasite of Cultured Turbot Scophthalmus maximus. Dis. Aquat. Org. 2003, 55, 133–144. [Google Scholar] [CrossRef]
- Ronza, P.; Robledo, D.; Bermudez, R.; Losada, A.P.; Pardo, B.G.; Sitja-Bobadilla, A.; Quiroga, M.I.; Martinez, P. RNA-Seq Analysis of Early Enteromyxosis in Turbot (Scophthalmus maximus): New Insights into Parasite Invasion and Immune Evasion Strategies. Int. J. Parasitol. 2016, 46, 507–517. [Google Scholar] [CrossRef]
- Filippi, M.-D. Mechanism of Diapedesis: Importance of the Transcellular Route. Adv. Immunol. 2016, 129, 25–53. [Google Scholar] [CrossRef]
- Ko, C.-Y.; Chang, W.-C.; Wang, J.-M. Biological Roles of CCAAT/Enhancer-Binding Protein Delta during Inflammation. J. Biomed. Sci. 2015, 22, 6. [Google Scholar] [CrossRef]
- Ramji, D.P.; Foka, P. CCAAT/Enhancer-Binding Proteins: Structure, Function and Regulation. Biochem. J. 2002, 365, 561–575. [Google Scholar] [CrossRef]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef]
- Arts, R.J.W.; Joosten, L.A.B.; Netea, M.G. Immunometabolic Circuits in Trained Immunity. Semin. Immunol. 2016, 28, 425–430. [Google Scholar] [CrossRef]
- Le Floc’h, N.; Melchior, D.; Obled, C. Modifications of Protein and Amino Acid Metabolism during Inflammation and Immune System Activation. Livest. Prod. Sci. 2004, 87, 37–45. [Google Scholar] [CrossRef]
- Kedia-Mehta, N.; Finlay, D.K. Competition for Nutrients and Its Role in Controlling Immune Responses. Nat. Commun. 2019, 10, 2123. [Google Scholar] [CrossRef]
- Masella, R.; Di Benedetto, R.; Varì, R.; Filesi, C.; Giovannini, C. Novel Mechanisms of Natural Antioxidant Compounds in Biological Systems: Involvement of Glutathione and Glutathione-Related Enzymes. J. Nutr. Biochem. 2005, 16, 577–586. [Google Scholar] [CrossRef]
- Torriglia, A.; Martin, E.; Jaadane, I. The Hidden Side of SERPINB1/Leukocyte Elastase Inhibitor. Semin. Cell Dev. Biol. 2017, 62, 178–186. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Murray, P.J. Cytokine Signaling Modules in Inflammatory Responses. Immunity 2008, 28, 477–487. [Google Scholar] [CrossRef]
- Nam, S.; Lim, J.-S. Essential Role of Interferon Regulatory Factor 4 (IRF4) in Immune Cell Development. Arch. Pharm. Res. 2016, 39, 1548–1555. [Google Scholar] [CrossRef]
- Purcell, M.K.; Smith, K.D.; Hood, L.; Winton, J.R.; Roach, J.C. Conservation of Toll-Like Receptor Signaling Pathways in Teleost Fish. Comp. Biochem. Physiol. Part D Genom. Proteom. 2006, 1, 77–88. [Google Scholar] [CrossRef]
- Piccinini, A.M.; Midwood, K.S. DAMPening Inflammation by Modulating TLR Signalling. Mediat. Inflamm. 2010, 2010. [Google Scholar] [CrossRef]
- Zou, J.; Secombes, C.J. Teleost Fish Interferons and Their Role in Immunity. Dev. Comp. Immunol. 2011, 35, 1376–1387. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, Y.-B.; Jiang, J.; Wang, B.; Chen, C.; Zhang, J.; Gui, J.-F. Gig1, a Novel Antiviral Effector Involved in Fish Interferon Response. Virology 2014, 448, 322–332. [Google Scholar] [CrossRef]
- Klamp, T.; Boehm, U.; Schenk, D.; Pfeffer, K.; Howard, J.C. A Giant GTPase, Very Large Inducible GTPase-1, Is Inducible by IFNs. J. Immunol. 2003, 171, 1255–1265. [Google Scholar] [CrossRef]
- Robledo, D.; Ronza, P.; Harrison, P.W.; Losada, A.P.; Bermúdez, R.; Pardo, B.G.; Redondo, M.J.; Sitjà-Bobadilla, A.; Quiroga, M.I.; Martínez, P. RNA-Seq Analysis Reveals Significant Transcriptome Changes in Turbot (Scophthalmus maximus) Suffering Severe Enteromyxosis. BMC Genom. 2014, 15, 1149. [Google Scholar] [CrossRef] [PubMed]
- Sitjà-Bobadilla, A.; Redondo, M.J.; Bermúdez, R.; Palenzuela, O.; Ferreiro, I.; Riaza, A.; Quiroga, I.; Nieto, J.M.; Álvarez-Pellitero, P. Innate and Adaptive Immune Responses of Turbot, Scophthalmus maximus (L.), Following Experimental Infection with Enteromyxum scophthalmi (Myxosporea: Myxozoa). Fish Shellfish Immunol. 2006, 21, 485–500. [Google Scholar] [CrossRef] [PubMed]
- Bermúdez, R.; Vigliano, F.; Marcaccini, A.; Sitjà-Bobadilla, A.; Quiroga, M.I.; Nieto, J.M. Response of Ig-Positive Cells to Enteromyxum scophthalmi (Myxozoa) Experimental Infection in Turbot, Scophthalmus maximus (L.): A Histopathological and Immunohistochemical Study. Fish Shellfish Immunol. 2006, 21, 501–512. [Google Scholar] [CrossRef] [PubMed]
| Gene Annotation | Mean TPM |
|---|---|
| beta-type globin | 224,710.66 |
| hemoglobin subunit alpha-1 | 215,830.97 |
| alpha-type globin | 107,927.82 |
| mhc class Ia chain | 22,249.08 |
| prostate stem cell antigen | 13,256.81 |
| golgin subfamily a member 5 | 6589.07 |
| mhc class Ia chain | 5201.32 |
| 40s ribosomal protein s30 | 4296.66 |
| ferritin, heavy subunit | 4142.98 |
| ribosomal protein s11 | 3925.73 |
| ribosomal protein, large, p0 | 3478.63 |
| elongation factor-1 alpha | 3273.46 |
| 40s ribosomal protein s20 isoform 2 | 3060.86 |
| beta-2-microglobulin | 3005.52 |
| mhc class Ia chain | 2821.07 |
| 40s ribosomal protein s14 | 2600.65 |
| 40s ribosomal protein s2 | 2514.28 |
| receptor for activated protein kinase c | 2422.57 |
| 60s ribosomal protein l21 | 2416.24 |
| ferritin, middle subunit | 2399.21 |
| elongation factor 2-like | 2350.56 |
| 5-aminolevulinate synthase, erythroid-specific, mitochondrial | 2211.96 |
| 40s ribosomal protein s9 | 2074.03 |
| ribosomal protein l6 | 2067.13 |
| muscle-specific beta 1 integrin binding protein 2 | 2052.37 |
| 60s ribosomal protein l13 | 2027.93 |
| cytochrome c oxidase subunit 1 | 1997.73 |
| elongation factor 1-alpha | 1944.72 |
| nudc domain-containing protein 2 | 1825.37 |
| aldehyde dehydrogenase family 16 member a1 | 1817.64 |
| 60s acidic ribosomal protein p1-like isoform x1 | 1798.65 |
| heat shock cognate 71 kda protein | 1798.03 |
| bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase | 1787.53 |
| 60s ribosomal protein l27 | 1745.03 |
| receptor expression-enhancing | 1735.88 |
| coxsackievirus and adenovirus receptor | 1729.71 |
| 60s ribosomal protein l17 | 1717.93 |
| tubulin beta-2c chain | 1695.68 |
| 40s ribosomal protein s23 | 1651.28 |
| immunoglobulin light chain | 1635.95 |
| trichohyalin-like | 1581.69 |
| 60s ribosomal protein l4-b | 1537.29 |
| thioredoxin-interacting protein | 1524.53 |
| 60s ribosomal protein l9 | 1521.54 |
| 60s ribosomal protein l8 | 1497.61 |
| myosin ic heavy chain-like | 1496.90 |
| tubulin beta-1 chain | 1476.30 |
| 40s ribosomal protein s13 | 1471.83 |
| 40s ribosomal protein s7 | 1410.55 |
| 60s ribosomal protein l5 | 1408.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ronza, P.; Álvarez-Dios, J.A.; Robledo, D.; Losada, A.P.; Romero, R.; Bermúdez, R.; Pardo, B.G.; Martínez, P.; Quiroga, M.I. Blood Transcriptomics of Turbot Scophthalmus maximus: A Tool for Health Monitoring and Disease Studies. Animals 2021, 11, 1296. https://doi.org/10.3390/ani11051296
Ronza P, Álvarez-Dios JA, Robledo D, Losada AP, Romero R, Bermúdez R, Pardo BG, Martínez P, Quiroga MI. Blood Transcriptomics of Turbot Scophthalmus maximus: A Tool for Health Monitoring and Disease Studies. Animals. 2021; 11(5):1296. https://doi.org/10.3390/ani11051296
Chicago/Turabian StyleRonza, Paolo, José Antonio Álvarez-Dios, Diego Robledo, Ana Paula Losada, Roberto Romero, Roberto Bermúdez, Belén G. Pardo, Paulino Martínez, and María Isabel Quiroga. 2021. "Blood Transcriptomics of Turbot Scophthalmus maximus: A Tool for Health Monitoring and Disease Studies" Animals 11, no. 5: 1296. https://doi.org/10.3390/ani11051296
APA StyleRonza, P., Álvarez-Dios, J. A., Robledo, D., Losada, A. P., Romero, R., Bermúdez, R., Pardo, B. G., Martínez, P., & Quiroga, M. I. (2021). Blood Transcriptomics of Turbot Scophthalmus maximus: A Tool for Health Monitoring and Disease Studies. Animals, 11(5), 1296. https://doi.org/10.3390/ani11051296








