Rule Discovery in Milk Content towards Mastitis Diagnosis: Dealing with Farm Heterogeneity over Multiple Years through Classification Based on Associations
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Generation of Year-Based Quartiled/Polynomial Datasets
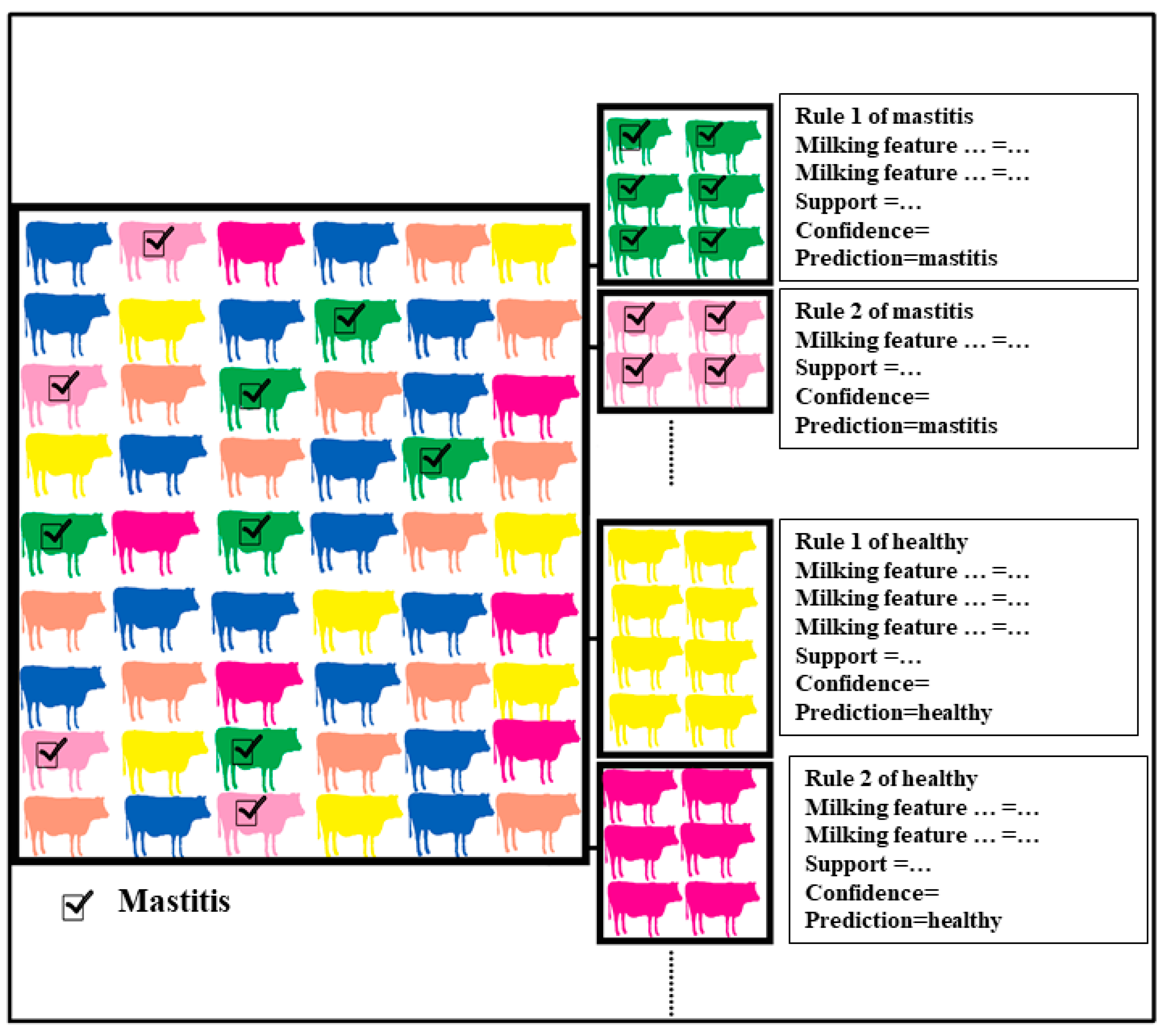
2.3. Rule Discovery in Scaled Data of Milking Features towards Subclinical Mastitis by Classification Based on Associations Model
2.4. Cross Validation to Estimate the Performance of the Developed Pipeline in Analysis of Future Unseen Data
2.5. Statistical Analysis
3. Results
3.1. Dealing with Farm Heterogeneity to Diagnose Subclinical Mastitis by Running Classification Based on Associations on the Ranked Datasets of Milking Features
- Coverage (Cov) %: The percentage of records containing the conditions in data set.
- Cover Count: The number of records containing the conditions.
- Support (Sup) %: The percentage of records containing both conditions and class.
- Support Count (SupCount): The number of records containing both conditions and class.
- Confidence (Conf) %: The ratio of support to coverage. This shows the precision of the rule in the data set.
3.2. Discovered Rules of Subclinical Mastitis Prediction from the Quartiled Milking Features in the First Year
- Rule 189: EC = Q4, Lact = Q1, Prot = Q4, Fat = Q4, Vol = Q1 -> class = Mastitis(Cover% = 0.24%, Conf% = 72.3%, CoverCount = 123, SupCount = 89, Sup% = 0.18%);
- Rule 190: Peak_Flow = Q1, EC = Q4, Lact = Q1, Prot = Q4, Fat = Q4 -> class = Mastitis(Cover% = 0.26%, Conf% = 69.8%, CoverCount = 129, SupCount = 90, Sup% = 0.18%);
- Rule 191: Peak_Flow = Q1, Milking_Time = Q1, EC = Q4, Lact = Q1, Fat= Q4, Vol = Q1 -> class = Mastitis(Cover% = 31%, Conf% = 69.4%, CoverCount = 157, SupCount = 109, Sup% = 0.22%);
- Rule 192: EC = Q4, Lact = Q1, Prot = Q4, Fat = Q4 -> class = MastitisCover% = 0.54%, Conf% = 67.66%, CoverCount = 269, SupCount = 182, Sup% = 0.36%);
- Rule 193: Peak_Flow = Q1, EC = Q4, Lact = Q1, Fat = Q4, Vol = Q1 -> class = Mastitis(Cover% = 0.64%, Conf% = 67.2%, CoverCount = 321, SupCount = 216, Sup% = 0.43%).
3.3. Rules of Subclinical Mastitis Prediction from the Quartiled Milking Features in the Second Year
- Rule 215: Peak_Flow = Q1, Milking_Time = Q1, EC = Q4, Lact = Q1, Prot = Q4, Vol = Q1 -> class = Mastitis(Cover% = 0.22%, Conf% = 77.3%, CoverCount = 110, SupCount = 85, Sup% = 0.17%);
- Rule 216: Peak_Flow = Q1, Milking_Time = Q3, EC = Q4, Lact = Q1, Fat= Q3, Vol = Q1 -> class = Mastitis(Cover% = 0.24%, Conf% = 73.96%, CoverCount = 119, SupCount = 88, Sup% = 0.18%);
- Rule 217: Peak_Flow = Q1, EC = Q4, Lact = Q1, Fat = Q4, Vol = Q1-> class = Mastitis(Cover% = 0.79%, Conf% = 72.5%, CoverCount = 397, SupCount = 288, Sup% = 0.58%);
- Rule 218: EC = Q4, Lact = Q1, Prot = Q4, Fat = Q4, Vol = Q1-> class = Mastitis(Cover% = 0.43%, Conf% = 69.3%, CoverCount = 215, SupCount = 149, Sup% = 0.30%);
- Rule 219: Peak_Flow = Q1, EC = Q4, Lact = Q1, Prot = Q4, Fat = Q4-> class = Mastitis(Cover% = 0.34%, Conf% = 69.2%, CoverCount = 169, SupCount = 117, Sup% = 0.23%).
3.4. Multiple Patterns Discovery towards Subclinical Mastitis Using the Quartiled Milking Features in the Third Year
- Rule 207: Peak_Flow = Q1, Milking_Time = Q2, EC = Q4, Lact = Q1, Fat = Q4-> class = Mastitis(Cover% = 0.32%, Conf% = 79.6%, CoverCount = 162, SupCount = 129, Sup% = 0.26%);
- Rule 208: Peak_Flow = Q1, EC = Q4, Lact = Q1, Fat = Q4, Vol = Q1-> class = Mastitis(Cover% = 0.78%, Conf% = 79.5%, CoverCount = 392, SupCount = 312, Sup% = 0.624%);
- Rule 209: Peak_Flow = Q1, Milking_Time = Q1, Lact = Q1, Fat = Q4, Vol = Q1-> class = Mastitis(Cover% = 0.47%, Conf% = 77.3%, CoverCount = 238, SupCount = 184, Sup% = 0.36%);
- Rule 210: Peak_Flow = Q1, EC = Q4, Lact = Q1, Prot = Q3, Vol = Q1-> class = Mastitis(Cover% = 0.43%, Conf% = 76.5%, CoverCount = 217, SupCount = 166, Sup% = 0.33%);
- Rule 211: Peak_Flow = Q1, Milking_Time = Q1, EC = Q4, Lact = Q1, Prot = Q1, Vol = Q1-> class = Mastitis(Cover% = 0.38%, Conf% = 76.17%, CoverCount = 193, SupCount = 147, Sup% = 0.29%);
- Rule 212: Peak_Flow = Q1, EC = Q4, Prot = Q4, Vol = Q1-> class = Mastitis(Cover% = 0.49%, Conf% = 75.4%, CoverCount = 248, SupCount = 187, Sup% = 0.37%).
3.5. Reproducible Rules of Subclinical Mastitis Diagnosis from Milking Features over Multiple Years
3.6. Feature Selection Using Classification Based on Associations
3.7. Risk Quartiles of Subclinical Mastitis Diagnosis
3.8. Ten-Fold cross Validation Analysis Showed Stablility of Classification Based on Associations-Derived Rules
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hogeveen, H.; Huijps, K.; Lam, T. Economic aspects of mastitis: New developments. N. Z. Vet. J. 2011, 59, 16–23. [Google Scholar] [CrossRef]
- Petrovski, K.; Buneski, G.; Trajcev, M. A review of the factors affecting the costs of bovine mastitis. J. S. Afr. Vet. Assoc. 2006, 77, 52–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pol, M.; Ruegg, P. Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. J. Dairy Sci. 2007, 90, 249–261. [Google Scholar] [CrossRef]
- Ebrahimie, E.; Ebrahimi, F.; Ebrahimi, M.; Tomlinson, S.; Petrovski, K.R. Hierarchical pattern recognition in milking parameters predicts mastitis prevalence. Comput. Electron. Agric. 2018, 147, 6–11. [Google Scholar] [CrossRef]
- Roy, S.; Vishwakarma, P.; Roy, M.; Sharma, M. Prevalence and control of bubaline mastitis in Chhattisgarh State in India. Pak. J. Zool. Suppl. Ser. 2009, 9, 281–287. [Google Scholar]
- Ebrahimi, M.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E.; Petrovski, K.R. Comprehensive analysis of machine learning models for prediction of sub-clinical mastitis: Deep learning and gradient-boosted trees outperform other models. Comput. Biol. Med. 2019, 114, 103456. [Google Scholar] [CrossRef]
- Bolstad, B.M.; Irizarry, R.A.; Åstrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Mohammadi-Dehcheshmeh, M.; Niazi, A.; Ebrahimi, M.; Tahsili, M.; Nurollah, Z.; Ebrahimi Khaksefid, R.; Ebrahimi, M.; Ebrahimie, E. Unified transcriptomic signature of arbuscular mycorrhiza colonization in roots of Medicago truncatula by integration of machine learning, promoter analysis, and direct merging meta-analysis. Front. Plant Sci. 2018, 9, 1550. [Google Scholar] [CrossRef] [Green Version]
- Sharifi, S.; Pakdel, A.; Ebrahimi, M.; Reecy, J.M.; Fazeli Farsani, S.; Ebrahimie, E. Integration of machine learning and meta-analysis identifies the transcriptomic bio-signature of mastitis disease in cattle. PLoS ONE 2018, 13, e0191227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helal, S.; Li, J.; Liu, L.; Ebrahimie, E.; Dawson, S.; Murray, D.J.; Long, Q. Predicting academic performance by considering student heterogeneity. Knowl. Based Syst. 2018, 161, 134–146. [Google Scholar] [CrossRef]
- Kargarfard, F.; Sami, A.; Ebrahimie, E. Knowledge discovery and sequence-based prediction of pandemic influenza using an integrated classification and association rule mining (CBA) algorithm. J. Biomed. Inform. 2015, 57, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Kargarfard, F.; Sami, A.; Mohammadi-Dehcheshmeh, M.; Ebrahimie, E. Novel approach for identification of influenza virus host range and zoonotic transmissible sequences by determination of host-related associative positions in viral genome segments. BMC Genom. 2016, 17, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kargarfard, F.; Sami, A.; Hemmatzadeh, F.; Ebrahimie, E. Identifying mutation positions in all segments of influenza genome enables better differentiation between pandemic and seasonal strains. Gene 2019, 697, 78–85. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Ma, Y.; Wong, C.-K. Classification using association rules: Weaknesses and enhancements. In Data Mining for Scientific and Engineering Applications; Springer: Berlin/Heidelberg, Germany, 2001; pp. 591–605. [Google Scholar]
- Chen, W.-C.; Hsu, C.-C.; Hsu, J.-N. Adjusting and generalizing CBA algorithm to handling class imbalance. Expert Syst. Appl. 2012, 39, 5907–5919. [Google Scholar] [CrossRef]
- Liu, B.; Hsu, W.; Ma, Y. Integrating classification and association rule mining. In Proceedings of the 4th International Conference on Discovery and Data Mining, New York, NY, USA, August 1998; pp. 80–86. [Google Scholar]
- Liu, B.; Hsu, W.; Ma, Y. Pruning and summarizing the discovered associations. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 15–18 August 1999; pp. 125–134. [Google Scholar]
- Liu, B.; Hsu, W.; Ma, Y. Mining association rules with multiple minimum supports. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 15–18 August 1999; pp. 337–341. [Google Scholar]
- Chen, W.-C.; Hsu, C.-C.; Chu, Y.-C. Increasing the effectiveness of associative classification in terms of class imbalance by using a novel pruning algorithm. Expert Syst. Appl. 2012, 39, 12841–12850. [Google Scholar] [CrossRef]
- Ebrahimie, E.; Ebrahimi, F.; Ebrahimi, M.; Tomlinson, S.; Petrovski, K.R. A large-scale study of indicators of sub-clinical mastitis in dairy cattle by attribute weighting analysis of milk composition features: Highlighting the predictive power of lactose and electrical conductivity. J. Dairy Res. 2018, 85, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Hahsler, M.; Johnson, I.; Kliegr, T.; Kucha, J. Associative Classification in R: Arc, arulesCBA, and rCBA. R J. 2019, 9, 254–267. [Google Scholar] [CrossRef]
- Neethirajan, S. The role of sensors, big data and machine learning in modern animal farming. Sens. Bio-Sens. Res. 2020, 29, 100367. [Google Scholar] [CrossRef]
- Cockburn, M. Application and Prospective Discussion of Machine Learning for the Management of Dairy Farms. Animals 2020, 10, 1690. [Google Scholar] [CrossRef] [PubMed]
- Anglart, D.; Hallén-Sandgren, C.; Emanuelson, U.; Rönnegård, L. Comparison of methods for predicting cow composite somatic cell counts. J. Dairy Sci. 2020, 103, 8433–8442. [Google Scholar] [CrossRef]
- Malcata, F.B.; Pepler, P.T.; O’Reilly, E.L.; Brady, N.; Eckersall, P.D.; Zadoks, R.N.; Viora, L. Point-of-care tests for bovine clinical mastitis: What do we have and what do we need? J. Dairy Res. 2020, 87, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Hyde, R.M.; Down, P.M.; Bradley, A.J.; Breen, J.E.; Hudson, C.; Leach, K.A.; Green, M.J. Automated prediction of mastitis infection patterns in dairy herds using machine learning. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Heide, E.; Veerkamp, R.; van Pelt, M.; Kamphuis, C.; Athanasiadis, I.; Ducro, B. Comparing regression, naive Bayes, and random forest methods in the prediction of individual survival to second lactation in Holstein cattle. J. Dairy Sci. 2019, 102, 9409–9421. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.; Aghalari, A.; Marufuzzaman, M.; Stone, A. Predicting dairy cattle heat stress using machine learning techniques. J. Dairy Sci. 2020, 104, 501–524. [Google Scholar] [CrossRef] [PubMed]
- Obayashi, T.; Kinoshita, K. Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res. 2009, 16, 249–260. [Google Scholar] [CrossRef] [Green Version]
- Adkins, P.R.; Middleton, J.R. Methods for diagnosing mastitis. Vet. Clin. Food Anim. Pract. 2018, 34, 479–491. [Google Scholar] [CrossRef]
- Costa, A.; Lopez-Villalobos, N.; Sneddon, N.; Shalloo, L.; Franzoi, M.; De Marchi, M.; Penasa, M. Invited review: Milk lactose—Current status and future challenges in dairy cattle. J. Dairy Sci. 2019, 102, 5883–5898. [Google Scholar] [CrossRef]
- Costa, A.; Neglia, G.; Campanile, G.; De Marchi, M. Milk somatic cell count and its relationship with milk yield and quality traits in Italian water buffaloes. J. Dairy Sci. 2020, 103, 5485–5494. [Google Scholar] [CrossRef]
- Costa, A.; Schwarzenbacher, H.; Mészáros, G.; Fuerst-Waltl, B.; Fuerst, C.; Sölkner, J.; Penasa, M. On the genomic regions associated with milk lactose in Fleckvieh cattle. J. Dairy Sci. 2019, 102, 10088–10099. [Google Scholar] [CrossRef]
- Stocco, G.; Pazzola, M.; Dettori, M.L.; Cipolat-Gotet, C.; Summer, A.; Vacca, G.M. Variation in caprine milk composition and coagulation as affected by udder health indicators. Int. Dairy J. 2019, 98, 9–16. [Google Scholar] [CrossRef]
- Helal, S.; Li, J.; Liu, L.; Ebrahimie, E.; Dawson, S.; Murray, D.J. Identifying key factors of student academic performance by subgroup discovery. Int. J. Data Sci. Anal. 2019, 7, 227–245. [Google Scholar] [CrossRef]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zecconi, A.; Vairani, D.; Cipolla, M.; Rizzi, N.; Zanini, L. Assessment of subclinical mastitis diagnostic accuracy by differential cell count in individual cow milk. Ital. J. Anim. Sci. 2019, 18, 460–465. [Google Scholar] [CrossRef]


| Year | Healthy Records | Subclinical Mastitis Records (Somatic Cell Count Cut-Off of ≥250,000 cells/mL) |
|---|---|---|
| Year 1 | 57,180 | 12,034 |
| Year 2 | 118,126 | 24,218 |
| Year 3 | 68,789 | 16,657 |
| Year | Mastitis Rules no. | Number and Percentage of Subclinical Mastitis-Detecting Rules across Years 1, 2, and 3. | ||||||
|---|---|---|---|---|---|---|---|---|
| Lactose_Q1 1 | EC_Q4 | Volume_Q1 | Peak_Flow_Q1 | Protein_Q4 | Fat_Q4 | Milking_Time_Q1 | ||
| Year 1 | 20 | 17 (85%) | 20 (100%) | 9 (45%) | 11 (55%) | 7 (35%) | 9 (45%) | 5 (25%) |
| Year 2 | 10 | 10 (100%) | 10 (100%) | 6 (60%) | 5 (50%) | 8 (80%) | 4(40%) | 2 (20%) |
| Year 3 | 35 | 33 (94.3%) | 18 (51.4%) | 24 (68.6%) | 15 (42.8%) | 9 (25.4%) | 14 (40%) | 10 (28.6%) |
| Ave. % ± SD | 93.1% ± 7.5% | 83.8% ± 28.1% | 57.9% ± 11.9% | 49.27% ± 6.1% | 46.8% ± 29.1% | 41.7% ± 2.9% | 24.5% ± 4.3% | |
| Year 1 | Year 2 | Year 3 |
|---|---|---|
| 17.9 | 16.6 | 19.7 |
| 17.1 | 17.1 | 19.8 |
| 17.8 | 17.0 | 19.5 |
| 17.9 | 17.4 | 19.9 |
| 17.1 | 16.7 | 19.1 |
| 17.7 | 17.1 | 18.9 |
| 17.1 | 17.2 | 19.0 |
| 17.8 | 16.5 | 19.6 |
| 17.5 | 17.4 | 18.5 |
| 16.7 | 16.5 | 20.6 |
| Avg 17.4 ± 0.4 | Avg 17.4 ± 0.4 | Avg 19.4 ± 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ebrahimie, E.; Mohammadi-Dehcheshmeh, M.; Laven, R.; Petrovski, K.R. Rule Discovery in Milk Content towards Mastitis Diagnosis: Dealing with Farm Heterogeneity over Multiple Years through Classification Based on Associations. Animals 2021, 11, 1638. https://doi.org/10.3390/ani11061638
Ebrahimie E, Mohammadi-Dehcheshmeh M, Laven R, Petrovski KR. Rule Discovery in Milk Content towards Mastitis Diagnosis: Dealing with Farm Heterogeneity over Multiple Years through Classification Based on Associations. Animals. 2021; 11(6):1638. https://doi.org/10.3390/ani11061638
Chicago/Turabian StyleEbrahimie, Esmaeil, Manijeh Mohammadi-Dehcheshmeh, Richard Laven, and Kiro Risto Petrovski. 2021. "Rule Discovery in Milk Content towards Mastitis Diagnosis: Dealing with Farm Heterogeneity over Multiple Years through Classification Based on Associations" Animals 11, no. 6: 1638. https://doi.org/10.3390/ani11061638
APA StyleEbrahimie, E., Mohammadi-Dehcheshmeh, M., Laven, R., & Petrovski, K. R. (2021). Rule Discovery in Milk Content towards Mastitis Diagnosis: Dealing with Farm Heterogeneity over Multiple Years through Classification Based on Associations. Animals, 11(6), 1638. https://doi.org/10.3390/ani11061638








