SARS-CoV-2, a Threat to Marine Mammals? A Study from Italian Seawaters
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Evaluation of the Susceptibility to SARS-CoV-2 of the Different Species of Marine Mammals in Mediterranean Sea through Comparative ACE-2 Receptor Analysis
2.1.1. ACE-2 Protein Sequences in Marine Mammals’ Analysis
2.1.2. Species Distribution along the Italian Coastline
2.1.3. Assessment of Species Susceptibility to SARS-CoV-2 Infection
2.1.4. Molecular Analyses (PCR and Sequencing)
2.1.5. Cross-Referencing of Conservation Status and Susceptibility
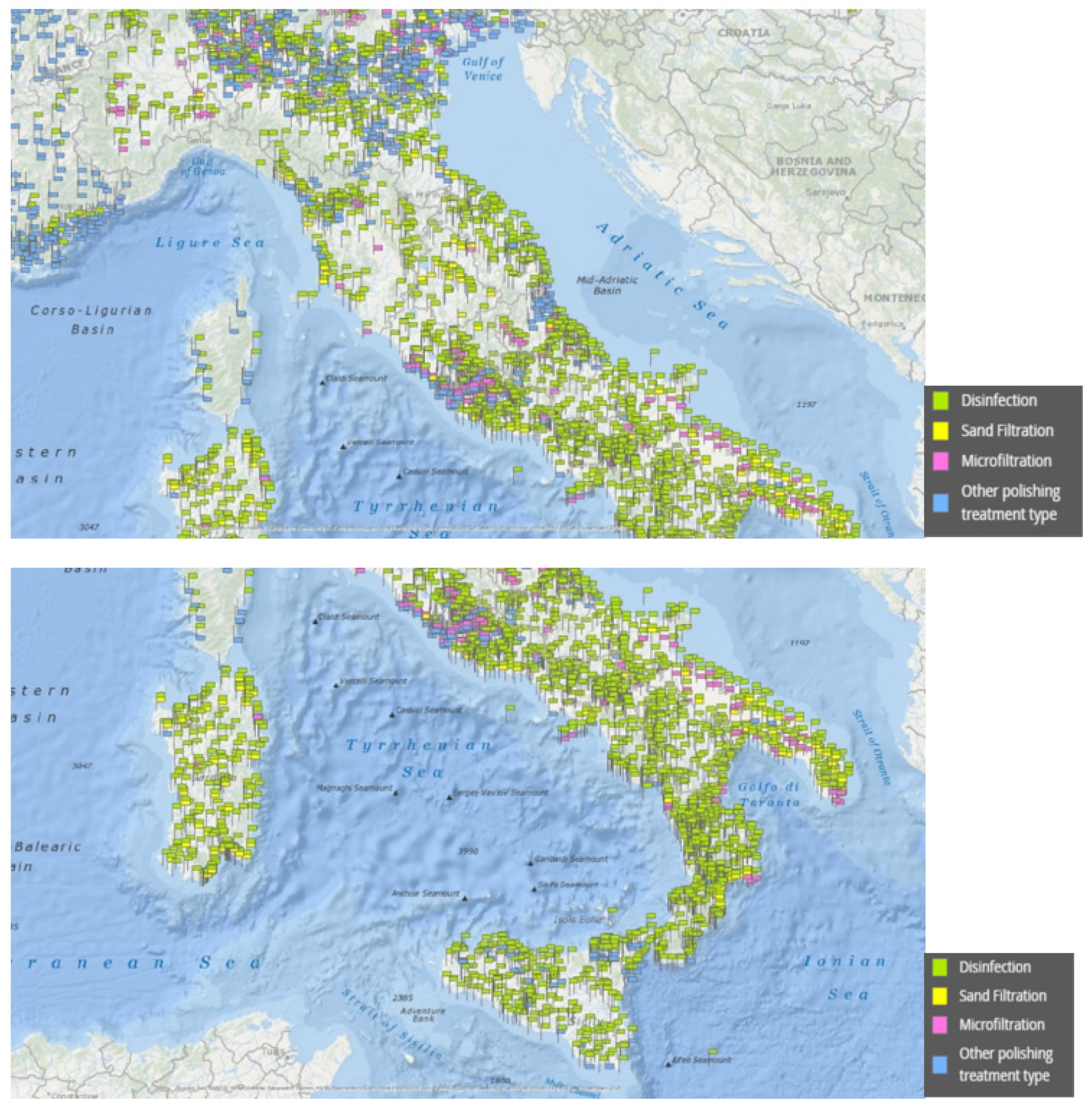
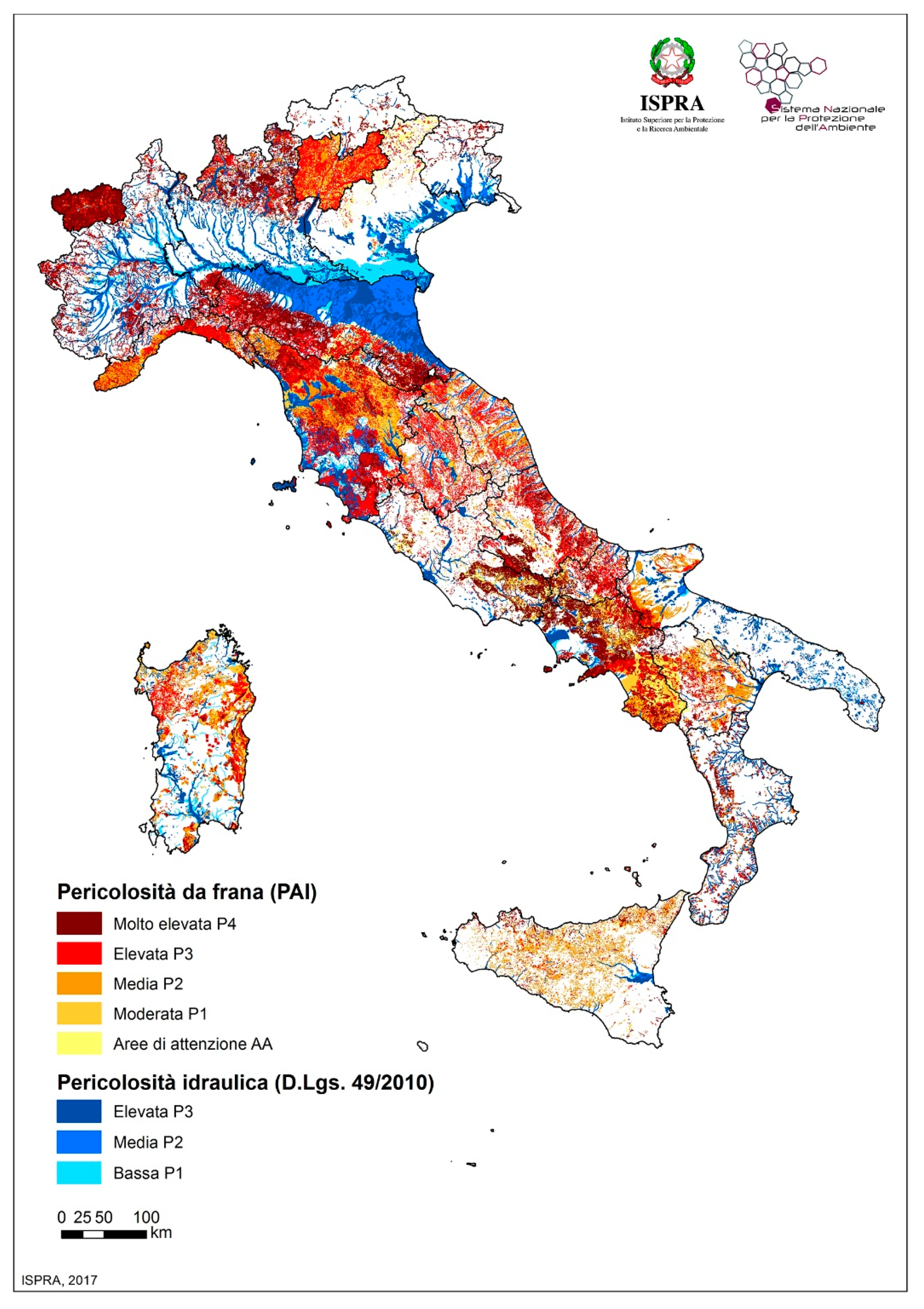
2.2. Geo-Mapping of Italian Wastewater Plants, Stretches of Coast and Species at Risk
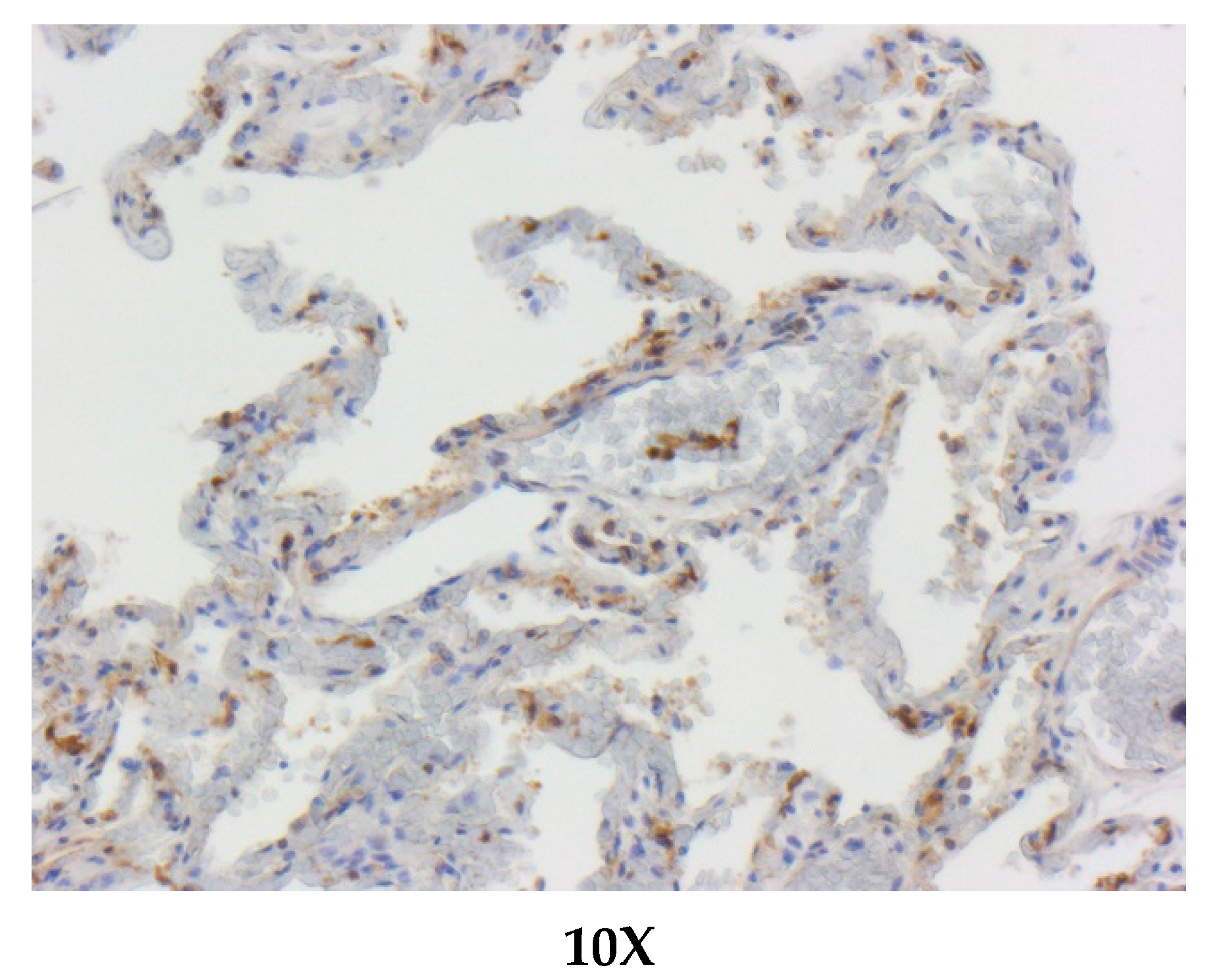
2.3. Immunohistochemistry
3. Results
3.1. Susceptibility of the Marine Mammal Species Examined to SARS-CoV-2
3.2. Wastewater Treatment Plants Conditions, Hydrogeological Vulnerability, and Risk to Marine Mammal Species in Italian Seas
3.3. Immunohistochemical (IHC) Characterization and Pulmonary Location of ACE-2
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A


References
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Ferraro, G.B.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhu, A.; Li, H.; Zheng, K.; Zhuang, Z.; Chen, Z.; Shi, Y.; Zhang, Z.; Chen, S.-B.; Liu, X.; et al. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020, 9, 991–993. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Sun, J.; Xu, Y.; Li, F.; Huang, X.; Li, H.; Zhao, J.; Huang, J.; Zhao, J. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020, 26, 1920–1922. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-W.; Li, J.-S.; Jin, M.; Zhen, B.; Kong, Q.-X.; Song, N.; Xiao, W.-J.; Yin, J.; Wei, W.; Wang, G.-J.; et al. Study on the resistance of severe acute respiratory syndrome-associated coronavirus. J. Virol. Methods 2005, 126, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.-S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.; Sarmah, A.K.; Chao, H.-P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2021, 193, 110265. [Google Scholar] [CrossRef]
- Collivignarelli, M.C.; Collivignarelli, C.; Miino, M.C.; Abbà, A.; Pedrazzani, R.; Bertanza, G. SARS-CoV-2 in sewer systems and connected facilities. Process. Saf. Environ. Prot. 2020, 143, 196–203. [Google Scholar] [CrossRef]
- Shutler, J.D.; Zaraska, K.; Holding, T.; Machnik, M.; Uppuluri, K.; Ashton, I.G.C.; Migdał, Ł.; Dahiya, R.S. Rapid assessment of SARS-CoV-2 transmission risk for fecally contaminated river water. ACS ES&T Water 2021, 1, 949–957. [Google Scholar] [CrossRef]
- Chin, A.W.H.; Chu, J.T.S.; A Perera, M.R.; Hui, K.P.Y.; Yen, H.-L.; Chan, M.C.W.; Peiris, M.; Poon, L.L.M. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 2020, 1, e10. [Google Scholar] [CrossRef]
- Wurtzer, S.; Marechal, V.; Mouchel, J.M.; Maday, Y.; Teyssou, R.; Richard, E.; Almayrac, J.L.; Moulin, L. Evaluation of lockdown impact on SARS-CoV-2 dynamics through viral genome quantification in paris wastewaters. MedRxiv 2020. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef]
- Rimoldi, S.G.; Stefani, F.; Gigantiello, A.; Polesello, S.; Comandatore, F.; Mileto, D.; Maresca, M.; Longobardi, C.; Mancon, A.; Romeri, F.; et al. Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Sci. Total Environ. 2020, 744, 140911. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Latorre, L.; Ballesteros, I.; Villacrés-Granda, I.; Granda, M.G.; Freire-Paspuel, B.; Ríos-Touma, B. SARS-CoV-2 in river water: Implications in low sanitation countries. Sci. Total Environ. 2020, 743, 140832. [Google Scholar] [CrossRef]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020, 737, 140405. [Google Scholar] [CrossRef] [PubMed]
- Gallus, W.A.; Parodi, A.; Maugeri, M. Possible impacts of a changing climate on intense Ligurian Sea rainfall events. Int. J. Clim. 2018, 38, e323–e329. [Google Scholar] [CrossRef]
- Funari, E.; Manganelli, M.; Sinisi, L. Impact of climate change on waterborne diseases. Annali dell’Istituto Superiore di Sanità 2012, 48, 473–487. [Google Scholar] [CrossRef]
- Griffin, D.W.; Donaldson, K.A.; Paul, J.H.; Rose, J.B. Pathogenic human viruses in coastal waters. Clin. Microbiol. Rev. 2003, 16, 129–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Guardo, G. Giovanni di guardo: Animal models and pathogenetic insights to Covid-19. J. Comp. Pathol. 2020, 179, e1. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Stoddart, A.K.; Gagnon, G.A.; Dellaire, G. Pandemic danger to the deep: The risk of marine mammals contracting SARS-CoV-2 from wastewater. Sci. Total Environ. 2021, 760, 143346. [Google Scholar] [CrossRef]
- Cao, Y.; Li, L.; Feng, Z.; Wan, S.; Huang, P.; Sun, X.; Wen, F.; Huang, X.; Ning, G.; Wang, W. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 2020, 6, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Zappulli, V.; Ferro, S.; Mazzariol, S.; Moccia, V.; Rensi, N.; Sammarco, A.; Torrigiani, F.; Verin, R.; Castagnaro, M.; Bonsembiante, F.; et al. Pathology of coronavirus infections: A review of lesions in animals in the one-health perspective. Animals 2020, 10, 2377. [Google Scholar] [CrossRef] [PubMed]
- Luan, J.; Jin, X.; Lu, Y.; Zhang, L. SARS-CoV-2 spike protein favors ACE2 from Bovidae and Cricetidae. J. Med. Virol. 2020, 92, 1649–1656. [Google Scholar] [CrossRef] [Green Version]
- Nabi, G.; Khan, S. Risk of COVID-19 pneumonia in aquatic mammals. Environ. Res. 2020, 188, 109732. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Hewson, I. Coronaviruses in the sea. Front. Microbiol. 2020, 11, 1795. [Google Scholar] [CrossRef]
- Albini, A.; Di Guardo, G.; Noonan, D.M.; Lombardo, M. The SARS-CoV-2 receptor, ACE-2, is expressed on many different cell types: Implications for ACE-inhibitor- and angiotensin II receptor blocker-based cardiovascular therapies. Intern. Emerg. Med. 2020, 15, 759–766. [Google Scholar] [CrossRef]
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.-P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Panigada, S.; Donovan, G.P.; Druon, J.-N.; Lauriano, G.; Pierantonio, N.; Pirotta, E.; Zanardelli, M.; Zerbini, A.N.; di Sciara, G.N. Satellite tagging of Mediterranean fin whales: Working towards the identification of critical habitats and the focussing of mitigation measures. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; Van Der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Wei, C.; Wan, L.; Yan, Q.; Wang, X.; Zhang, J.; Yang, X.; Zhang, Y.; Fan, C.; Li, D.; Deng, Y.; et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020, 2, 1391–1400. [Google Scholar] [CrossRef] [PubMed]
- Fossi, M.; Marsili, L.; Lauriano, G.; Fortuna, C.; Canese, S.; Ancora, S.; Leonzio, C.; Romeo, T.; Merino, R.; Abad, E.; et al. Assessment of toxicological status of a SW Mediterranean segment population of striped dolphin (Stenella coeruleoalba) using skin biopsy. Mar. Environ. Res. 2004, 58, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Cózar, A.; Sanz-Martín, M.; Martí, E.; González-Gordillo, J.I.; Ubeda, B.; Gálvez, J. Ángel; Irigoien, X.; Duarte, C.M. Plastic Accumulation in the Mediterranean Sea. PLoS ONE 2015, 10, e0121762. [Google Scholar] [CrossRef] [Green Version]
- Panigada, S.; Pesante, G.; Zanardelli, M.; Capoulade, F.; Gannier, A.; Weinrich, M.T. Mediterranean fin whales at risk from fatal ship strikes. Mar. Pollut. Bull. 2006, 52, 1287–1298. [Google Scholar] [CrossRef]
- Di Guardo, G.; Mazzariol, S. Dolphin Morbillivirus: A lethal but valuable infection model. Emerg. Microbes Infect. 2013, 2, 1–11. [Google Scholar] [CrossRef]
- Bossard, G. Marine mammals as sentinel species for oceans and human health. Oceanography 2006, 19, 134–137. [Google Scholar] [CrossRef] [Green Version]
- Van Bressem, M.-F.; Duignan, P.J.; Banyard, A.; Barbieri, M.; Colegrove, K.M.; De Guise, S.; Di Guardo, G.; Dobson, A.; Domingo, M.; Fauquier, D.; et al. Cetacean morbillivirus: Current knowledge and future directions. Viruses 2014, 6, 5145–5181. [Google Scholar] [CrossRef]
- Duignan, P.J.; Van Bressem, M.-F.; Baker, J.D.; Barbieri, M.; Colegrove, K.M.; De Guise, S.; De Swart, R.L.; Di Guardo, G.; Dobson, A.; Duprex, W.P.; et al. Phocine distemper virus: Current knowledge and future directions. Viruses 2014, 6, 5093–5134. [Google Scholar] [CrossRef] [PubMed]
- Di Guardo, G.; Mazzariol, S. Cetacean morbillivirus: A Land-to-Sea Journey and Back? Virol. Sin. 2019, 34, 240–242. [Google Scholar] [CrossRef] [PubMed]
- Mathavarajah, S.; Dellaire, G. Lions, tigers and kittens too: ACE2 and susceptibility to COVID-19. Evol. Med. Public Health 2020, 2020, 109–113. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020, 94, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abassi, Z.; Knaney, Y.; Karram, T.; Heyman, S.N. The lung macrophage in SARS-CoV-2 infection: A friend or a foe? Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Grattarola, C.; Gallina, S.; Giorda, F.; Pautasso, A.; Ballardini, M.; Iulini, B.; Varello, K.; Goria, M.; Peletto, S.; Masoero, L.; et al. First report of Salmonella 1,4,[5],12:i:- in free-ranging striped dolphins (Stenella coeruleoalba), Italy. Sci. Rep. 2019, 9, 6061. [Google Scholar] [CrossRef] [Green Version]
- Grattarola, C.; Giorda, F.; Iulini, B.; Pintore, M.D.; Pautasso, A.; Zoppi, S.; Goria, M.; Romano, A.; Peletto, S.; Varello, K.; et al. Meningoencephalitis and Listeria monocytogenes, Toxoplasma gondii and Brucella spp. coinfection in a dolphin in Italy. Dis. Aquat. Org. 2016, 118, 169–174. [Google Scholar] [CrossRef]
- Decaro, N.; Lorusso, A. Novel human coronavirus (SARS-CoV-2): A lesson from animal coronaviruses. Veter. Microbiol. 2020, 244, 108693. [Google Scholar] [CrossRef] [PubMed]
- Mihindukulasuriya, K.A.; Wu, G.; Leger, J.S.; Nordhausen, R.W.; Wang, D. Identification of a novel coronavirus from a beluga whale by using a panviral microarray. J. Virol. 2008, 82, 5084–5088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reeves, R.; Notarbartolo di Sciara, G. The Status and Distribution of Cetaceans in the Black Sea and Mediterranean Sea; IUCN Centre for Mediterranean Cooperation: Málaga, Spain, 2006; p. 137. [Google Scholar]
- Bearzi, G.; Holcer, D.; Di Sciara, G.N. The role of historical dolphin takes and habitat degradation in shaping the present status of northern Adriatic cetaceans. Aquat. Conserv. Mar. Freshw. Ecosyst. 2004, 14, 363–379. [Google Scholar] [CrossRef]
- Lauriano, G.; Pierantonio, N.; Donovan, G.; Panigada, S. Abundance and distribution of tursiops truncatus in the western mediterranean sea: An assessment towards the marine strategy framework directive requirements. Mar. Environ. Res. 2014, 100, 86–93. [Google Scholar] [CrossRef]
- Gnone, G.; Bellingeri, M.; Dhermain, F.; Dupraz, F.; Nuti, S.; Bedocchi, D.; Moulins, A.; Rosso, M.; Alessi, J.; McCrea, R.S.; et al. Distribution, abundance, and movements of the bottlenose dolphin (Tursiops truncatus) in the Pelagos Sanctuary MPA (north-west Mediterranean Sea). Aquat. Conserv. Mar. Freshw. Ecosyst. 2011, 21, 372–388. [Google Scholar] [CrossRef]
- Aguilar, A. Population biology, conservation threats and status of Mediterranean striped dolphins (Stenella coeruleo-alba). J. Cetacean Res. 2000, 2, 17–26. [Google Scholar]
- Di Sciara, G.N.; Hoyt, E.; Reeves, R.; Ardron, J.; Marsh, H.; Vongraven, D.; Barr, B. Place-based approaches to marine mammal conservation. Aquat. Conserv. Mar. Freshw. Ecosyst. 2016, 26, 85–100. [Google Scholar] [CrossRef]
- Canese, S.; Cardinali, A.; Fortuna, C.M.; Giusti, M.; Lauriano, G.; Salvati, E.; Greco, S. The first identified winter feeding ground of fin whales (Balaenoptera physalus) in the Mediterranean Sea. J. Mar. Biol. Assoc. UK 2006, 86, 903–907. [Google Scholar] [CrossRef]
- Bearzi, G.; Reeves, R.R.; di Sciara, G.N.; Politi, E.; Canadas, A.N.A.; Frantzis, A.; Mussi, B. Ecology, status and conservation of short-beaked common dolphins Delphinus delphis in the Mediterranean Sea. Mammal Rev. 2003, 33, 224–252. [Google Scholar] [CrossRef] [Green Version]
- Mazzariol, S.; Centelleghe, C.; Beffagna, G.; Povinelli, M.; Terracciano, G.; Cocumelli, C.; Pintore, A.; Denurra, D.; Casalone, C.; Pautasso, A.; et al. Mediterranean fin whales (Balaenoptera physalus) threatened by dolphin morbillivirus. Emerg. Infect. Dis. 2015, 22, 302–305. [Google Scholar] [CrossRef]
- Esteban, R.; Verborgh, P.; Gauffier, P.; Alarcón, D.; Salazar-Sierra, J.; Giménez, J.; Foote, A.; de Stephanis, R. Conservation status of killer whales, orcinus orca, in the strait of gibraltar. Adv. Mar. Biol. 2016, 75, 141–172. [Google Scholar] [CrossRef] [PubMed]


| Present Regularly | Occasional |
|---|---|
| Cetacea | |
| Balaenoptera physalus | Balaenoptera acutorostrata |
| Physeter macrocephalus | Pseudorca crassidens |
| Ziphius cavirostris | Megaptera novaengliae |
| Globicephala melas | |
| Orcinus orca | |
| Grampus griseus | |
| Tursiops truncates | |
| Stenella coeruleoalba | |
| Delphinus delphis | |
| Delphinus capensis | |
| Steno bredanensis | |
| Pinnipedia | |
| Monachus monachus | |
| Primer | Nucleotide Sequence (5′-3′) | Amplicon Size (bp) |
|---|---|---|
| CetACE2_19-90 F | CTCCTTCTCAGCCTTGTTGC | 285 bp |
| CetACE2_19-90 R | CTGAAGGRCCTGCAATTGAC | |
| CetACE2_322-393 F | AGTCCTGGGATGCAAAGAGG | 492 bp |
| CetACE2_322-393 R | AGACCATTCYCCTCCACTTT |
| Sequence Position Number | 19 | 24 | 27 | 28 | 30 | 31 | 34 | 35 | 37 | 38 | 41 | 42 | 45 | 53 | 79 | 82 | 83 | 90 | 322 | 330 | 353 | 354 | 355 | 357 | 393 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Homo sapiens | S | Q | T | F | D | K | H | E | E | D | Y | Q | L | N | L | M | Y | N | N | N | K | G | D | R | R |
| Balaenoptera physalus | S | Q | T | F | Q | K | H | E | E | D | H | E | L | N | I | T | Y | N | N | N | K | G | D | R | R |
| Physeter macrocephalus | S | Q | T | F | Q | K | H | E | E | D | Y | Q | L | N | T | T | Y | N | N | N | K | G | D | R | R |
| Ziphius cavirostris | P | K | T | F | Q | K | H | E | E | D | Y | Q | L | N | T | T | Y | N | N | N | K | G | D | R | R |
| Globicephala melas | S | R | T | F | Q | K | R | E | E | D | Y | Q | L | N | I | T | Y | N | N | N | K | G | D | R | R |
| Tursiops truncatus | S | R | T | F | Q | K | R | E | E | D | Y | Q | L | N | I | T | Y | N | N | N | K | G | D | R | R |
| Stenella coeruleoalba | S | R | T | F | Q | K | R | E | E | D | Y | Q | L | N | I | T | Y | N | N | N | K | G | D | R | R |
| Balaenoptera acutorostrata | S | Q | T | F | Q | K | H | E | E | D | Y | R | L | N | I | T | Y | N | N | N | K | G | D | R | R |
| Orcinus orca | S | R | T | F | Q | K | R | E | E | D | Y | Q | L | N | I | T | Y | N | N | N | K | G | D | R | R |
| Megaptera novaengliae | S | Q | T | F | Q | K | R | E | E | D | Y | E | . | N | . | T | Y | N | N | N | K | G | D | R | R |
| Color legend:. | |||||||||||||||||||||||||
| Non-conservative replacement | |||||||||||||||||||||||||
| Conservative replacement | |||||||||||||||||||||||||
| Species | IUCN Status |
|---|---|
| Balaenoptera acutorostrata (Minke whale) | LC (gl, 2018) |
| Megaptera novaeangliae (Humpbeak whale) | LC (gl, 2018) |
| Orcinus orca (Killer whale) | CE (Strait of Gibraltar subpopulation, 2019) DD (gl, 2017) |
| Pseudorca crassidens (False killer whale) | NT (gl, 2018) |
| Steno bredanensis (rough-toothed dolphin) | LC (gl, 2018) |
| Delphinus delphis (short beaked common dolphin) | EN (it, 2013), EN (med, 2003), LC (gl, 2008) |
| Delphinus capensis (long beaked common dolphin) | EN (it, 2013), EN (med, 2003), LC (gl, 2008) |
| Physeter macrocephalus (Sperm whale) | EN (it, 2013), EN (med, 2006), VU (gl, 2008) |
| Balaenoptera physalus (Fin whale) | VU (it, 2013), VU (med, 2010), VU (gl, 2018) |
| Tursiops truncatus (Common bottlenose dolphin) | NT (it, 2013), VU (med, 2009), LC (gl, 2018) |
| Stenella coeruleoalba (Striped dolphin) | LC (it, 2013), VU (med, 2010), LC (gl, 2018) |
| Globicephala melas (Pilot whale) | DD (it, 2013), DD (med, 2010), LC (gl, 2018) |
| Grampus griseus (Risso’s dolphin) | DD (it, 2013), DD (med, 2010), LC (gl, 2018) |
| Ziphius cavirostris (Cuvier’s beaked whale) | DD (it, 2013), DD (med, 2006), LC (gl, 2008) |
| Monachus monachus | DD (it, 2013), CR (med, 2008), EN (gl, 2015) |
| Species | IHC Results—LUNG Samples |
|---|---|
| Ziphius cavirostris | bronchiolar epithelial cells and alveolar macrophages + |
| Tursiops truncatus | bronchiolar epithelial cells + |
| Stenella coeruleoalba | bronchiolar epithelial cells and alveolar macrophages + |
| Grampus griseus | bronchiolar epithelial cells + |
| Pseudorca crassidens | bronchiolar epithelial cells and alveolar macrophages + |
| Balenoptera physalus | bronchiolar epithelial cells + |
| Delphinus delphis | bronchiolar epithelial cells + |
| Order | Species | IUCN Status | No. Non-Conservative Replacements | No. Residues Identical to Human | Mutations * | Predicted Susceptibility to SARS-CoV 2 Based on Amino Acidy Similarity | |
|---|---|---|---|---|---|---|---|
| Regular | Cetacea | B. physalus | VU (med, 2010) (VU it, 2013) | 1 | 20 | D30Q, Y41H, Q42E, L79I, M82T | High |
| Cetacea | P. macrocephalus | EN (med, 2006) (EN it, 2013) | 2 | 22 | D30Q, L79T, M82T | Medium | |
| Cetacea | Z. cavirostris | DD (med, 2006) (DD it, 2013) | 2 | 20 | S19P, Q24K, D30Q, L79T, M82T | Medium | |
| Cetacea | G. melas | DD (med, 2010) (DD it, 2013) | 1 | 20 | Q24R, D30Q, H34R, L79I, M82T | High | |
| Cetacea | T. truncatus | VU (med, 2009) (NT it, 2013) | 1 | 20 | Q24R, D30Q, H34R, L79I, M82T | High | |
| Cetacea | Orcinus orca | DD (med, 2019) | 1 | 20 | Q24R, D30Q, H34R, L79I, M82T | High | |
| Cetacea | S. coeruleoalba | VU (med, 2010) (LC it, 2013) | 1 | 20 | Q24R, D30Q, H34R, L79I, M82T | High | |
| Irregular | Cetacea | B. acutorostrata | LC (gl 2018) | 1 | 21 | D30Q, Q42R, L79I, M82T | High |
| Cetacea | M. novaengliae | LC (gl. 2018) | 1 | 21 | D30Q, H34R, Q42E, M82T | High |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Audino, T.; Grattarola, C.; Centelleghe, C.; Peletto, S.; Giorda, F.; Florio, C.L.; Caramelli, M.; Bozzetta, E.; Mazzariol, S.; Di Guardo, G.; et al. SARS-CoV-2, a Threat to Marine Mammals? A Study from Italian Seawaters. Animals 2021, 11, 1663. https://doi.org/10.3390/ani11061663
Audino T, Grattarola C, Centelleghe C, Peletto S, Giorda F, Florio CL, Caramelli M, Bozzetta E, Mazzariol S, Di Guardo G, et al. SARS-CoV-2, a Threat to Marine Mammals? A Study from Italian Seawaters. Animals. 2021; 11(6):1663. https://doi.org/10.3390/ani11061663
Chicago/Turabian StyleAudino, Tania, Carla Grattarola, Cinzia Centelleghe, Simone Peletto, Federica Giorda, Caterina Lucia Florio, Maria Caramelli, Elena Bozzetta, Sandro Mazzariol, Giovanni Di Guardo, and et al. 2021. "SARS-CoV-2, a Threat to Marine Mammals? A Study from Italian Seawaters" Animals 11, no. 6: 1663. https://doi.org/10.3390/ani11061663
APA StyleAudino, T., Grattarola, C., Centelleghe, C., Peletto, S., Giorda, F., Florio, C. L., Caramelli, M., Bozzetta, E., Mazzariol, S., Di Guardo, G., Lauriano, G., & Casalone, C. (2021). SARS-CoV-2, a Threat to Marine Mammals? A Study from Italian Seawaters. Animals, 11(6), 1663. https://doi.org/10.3390/ani11061663









