Characterization of Extended-Spectrum Cephalosporin (ESC) Resistance in Salmonella Isolated from Chicken and Identification of High Frequency Transfer of blaCMY-2 Gene Harboring Plasmid In Vitro and In Vivo
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Antimicrobial Susceptibility Test
2.3. Identification of β-Lactamases
2.4. Plasmid Replicon Typing
2.5. In Vitro Conjugation Experiment
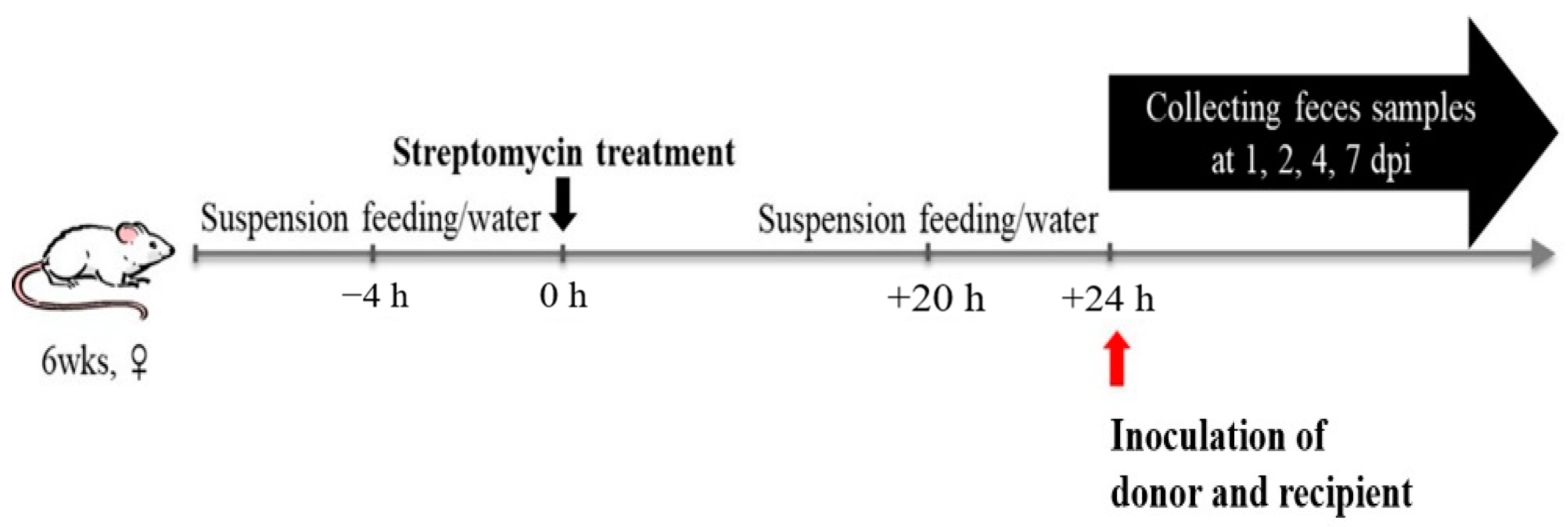
2.6. In Vivo Transfer Experiment
2.7. Competition Experiment In Vitro
3. Results
3.1. Characterization of Bacterial Strains
3.2. In Vitro Transfer
3.3. In Vivo Transfer
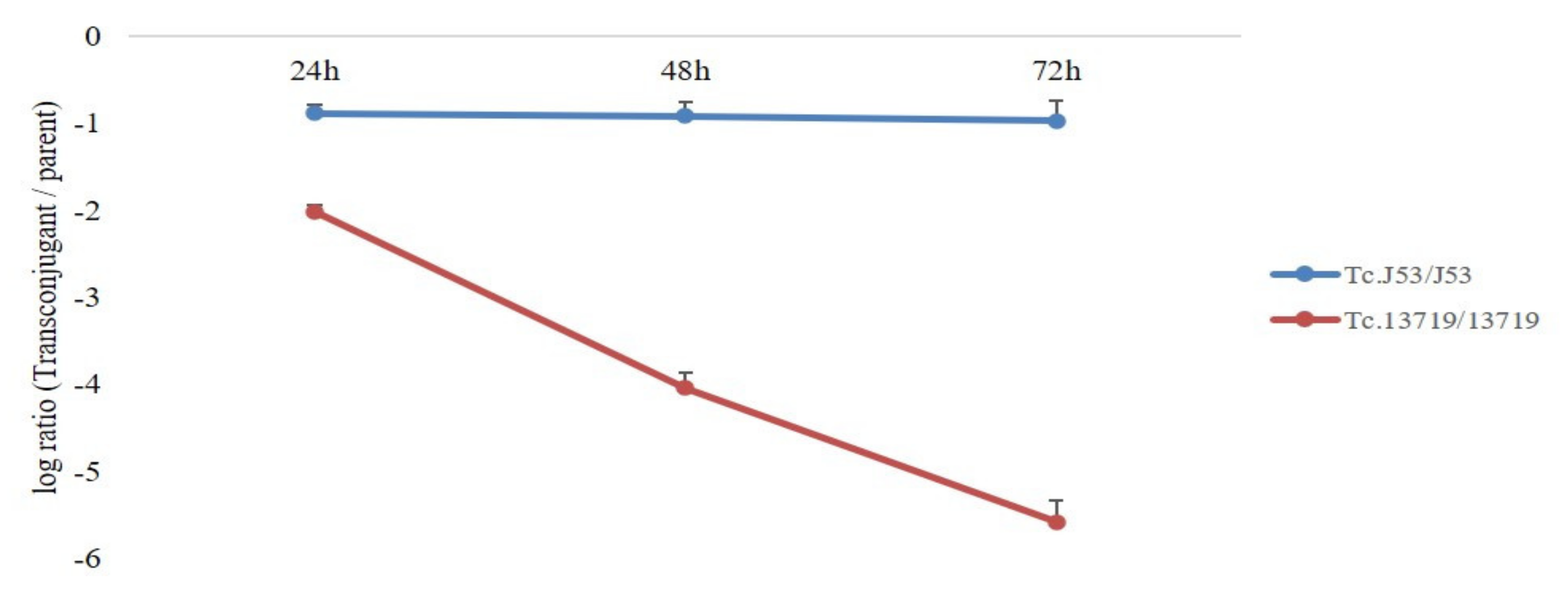
3.4. Fitness Cost Assessment by Competition Experiment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Mo, S.S.; Sunde, M.; Ilag, H.K.; Langsrud, S.; Heir, E. Transfer Potential of Plasmids Conferring Extended-Spectrum-Cephalosporin Resistance in Escherichia coli from Poultry. Appl. Environ. Microbiol. 2017, 83, 83. [Google Scholar] [CrossRef] [PubMed]
- Doublet, B.; Praud, K.; Nguyen-Ho-Bao, T.; Argudín, M.A.; Bertrand, S.; Butaye, P.; Cloeckaert, A. Extended-spectrum -lactamase- and AmpC -lactamase-producing D-tartrate-positive Salmonella enterica serovar Paratyphi B from broilers and human patients in Belgium, 2008–2010. J. Antimicrob. Chemother. 2013, 69, 1257–1264. [Google Scholar] [CrossRef] [PubMed]
- Na, S.H.; Moon, D.C.; Kang, H.Y.; Song, H.-J.; Kim, S.-J.; Choi, J.-H.; Yoon, J.W.; Yoon, S.-S.; Lim, S.-K. Molecular characteristics of extended-spectrum β-lactamase/AmpC-producing Salmonella enterica serovar Virchow isolated from food-producing animals during 2010–2017 in South Korea. Int. J. Food Microbiol. 2020, 322, 108572. [Google Scholar] [CrossRef]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Depoorter, P.; Persoons, D.; Uyttendaele, M.; Butaye, P.; de Zutter, L.; Dierick, K.; Herman, L.; Imberechts, H.; van Huffel, X.; Dewulf, J. Assessment of human exposure to 3rd generation cephalosporin resistant E. coli (CREC) through consumption of broiler meat in Belgium. Int. J. Food Microbiol. 2012, 159, 30–38. [Google Scholar] [CrossRef]
- Gama, J.A.; Zilhão, R.; Dionisio, F. Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: Plasmids promote the immigration of other plasmids but repress co-colonizing plasmids. Plasmid 2017, 93, 6–16. [Google Scholar] [CrossRef]
- Dionisio, F.; Conceição, I.; Marques, A.C.; Fernandes, L.; Gordo, I. The evolution of a conjugative plasmid and its ability to increase bacterial fitness. Biol. Lett. 2005, 1, 250–252. [Google Scholar] [CrossRef]
- Sommer, M.O.A.; Dantas, G.; Church, G.M. Functional Characterization of the Antibiotic Resistance Reservoir in the Human Microflora. Science 2009, 325, 1128–1131. [Google Scholar] [CrossRef]
- Shang, K.; Wei, B.; Kang, M. Distribution and dissemination of antimicrobial-resistant Salmonella in broiler farms with or without enrofloxacin use. BMC Vet. Res. 2018, 14, 1–14. [Google Scholar] [CrossRef]
- Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; CLSI Supplement M100; CLSI: Wayne, PA, USA, 2017. [Google Scholar]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters; European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2019. [Google Scholar]
- National Antimicrobial Resistance Monitoring System (NARMS). National Antimicrobial Resistance Monitoring System for Enteric Bacteria: Human Isolates Surveillance Report for 2015 (Final Report); Department of Health and Human Services, CDC: Atlanta, GA, USA, 2018.
- Dallenne, C.; da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Pérez, F.J.; Hanson, N.D. Detection of Plasmid-Mediated AmpC -Lactamase Genes in Clinical Isolates by Using Multiplex PCR. J. Clin. Microbiol. 2002, 40, 2153–2162. [Google Scholar] [CrossRef] [PubMed]
- Mata, C.; Navarro, F.; Miró, E.; Walsh, T.; Mirelis, B.; Toleman, M. Prevalence of SXT/R391-like integrative and conjugative elements carrying blaCMY-2 in Proteus mirabilis. J. Antimicrob. Chemother. 2011, 66, 2266–2270. [Google Scholar] [CrossRef] [PubMed]
- Barletta, F.; Mercado, E.H.; Lluque, A.; Ruiz, J.; Cleary, T.G.; Ochoa, T.J. Multiplex Real-Time PCR for Detection of Campylobacter, Salmonella, and Shigella. J. Clin. Microbiol. 2013, 51, 2822–2829. [Google Scholar] [CrossRef]
- Barthel, M.; Hapfelmeier, S.; Quintanilla-Martínez, L.; Kremer, M.; Rohde, M.; Hogardt, M.; Pfeffer, K.; Rüssmann, H.; Hardt, W.-D. Pretreatment of Mice with Streptomycin Provides a Salmonella enterica Serovar Typhimurium Colitis Model That Allows Analysis of Both Pathogen and Host. Infect. Immun. 2003, 71, 2839–2858. [Google Scholar] [CrossRef]
- Göttig, S.; Riedel-Christ, S.; Saleh, A.; Kempf, V.A.; Hamprecht, A. Impact of blaNDM-1 on fitness and pathogenicity of Escherichia coli and Klebsiella pneumoniae. Int. J. Antimicrob. Agents 2016, 47, 430–435. [Google Scholar] [CrossRef]
- Bates, S.; Cashmore, A.M.; Wilkins, B.M. IncP Plasmids Are Unusually Effective in Mediating Conjugation of Escherichia coli and Saccharomyces cerevisiae: Involvement of the Tra2 Mating System. J. Bacteriol. 1998, 180, 6538–6543. [Google Scholar] [CrossRef] [PubMed]
- Lampkowska, J.; Feld, L.; Monaghan, A.; Toomey, N.; Schjørring, S.; Jacobsen, B.; van der Voet, H.; Andersen, S.R.; Bolton, D.; Aarts, H.; et al. A standardized conjugation protocol to asses antibiotic resistance transfer between lactococcal species. Int. J. Food Microbiol. 2008, 127, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Thingholm, K.R.; Hertz, F.B.; Løbner-Olesen, A.; Frimodt-Møller, N.; Nielsen, K.L. Escherichia coli belonging to ST131 rarely transfers blactx-m-15 to fecal Escherichia coli. Infect. Drug Resist. 2019, 12, 2429–2435. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A. Plasmids in Gram negatives: Molecular typing of resistance plasmids. Int. J. Med. Microbiol. 2011, 301, 654–658. [Google Scholar] [CrossRef]
- Bauernfeind, A.; Stemplinger, I.; Jungwirth, R.; Giamarellou, H. Characterization of the plasmidic beta-lactamase CMY-2, which is responsible for cephamycin resistance. Antimicrob. Agents Chemother. 1996, 40, 221–224. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, S.; White, D.G.; Schroeder, C.M.; Lu, R.; Yang, H.; McDermott, P.F.; Ayers, S.; Meng, J. Characterization of Multiple-Antimicrobial-Resistant Salmonella Serovars Isolated from Retail Meats. Appl. Environ. Microbiol. 2004, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Schjørring, S.; Struve, C.; Krogfelt, K.A. Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J. Antimicrob. Chemother. 2008, 62, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Lalruatdiki, A.; Dutta, T.K.; Roychoudhury, P.; Subudhi, P.K. Extended-spectrum β-lactamases producing multidrug resistance Escherichia coli, Salmonella and Klebsiella pneumoniae in pig population of Assam and Meghalaya, India. Vet. World 2018, 11, 868–873. [Google Scholar] [CrossRef]
- Faure, S.; Perrin-Guyomard, A.; Delmas, J.-M.; Laurentie, M. Impact of Therapeutic Treatment with β-Lactam on Transfer of the blaCTX-M-9 Resistance Gene from Salmonella enterica Serovar Virchow to Escherichia coli in Gnotobiotic Rats. Appl. Environ. Microbiol. 2009, 75, 5523–5528. [Google Scholar] [CrossRef] [PubMed]
- Dahl, K.H.; Mater, D.D.G.; Flores, M.J.; Johnsen, P.J.; Midtvedt, T.; Corthier, G.; Sundsfjord, A. Transfer of plasmid and chromosomal glycopeptide resistance determinants occurs more readily in the digestive tract of mice than in vitro and exconjugants can persist stably In Vivo in the absence of glycopeptide selection. J. Antimicrob. Chemother. 2007, 59, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Licht, T.R.; Christensen, B.; Krogfelt, K.A.; Molin, S. Plasmid transfer in the animal intestine and other dynamic bacterial populations: The role of community structure and environment. Microbiology 1999, 145, 2615–2622. [Google Scholar] [CrossRef]
- Su, L.-H.; Chiu, C.-H.; Chu, C.; Wang, M.-H.; Chia, J.-H.; Wu, T.-L. In Vivo Acquisition of Ceftriaxone Resistance in Salmonella enterica Serotype Anatum. Antimicrob. Agents Chemother. 2003, 47, 563–567. [Google Scholar] [CrossRef][Green Version]
- Sunde, M. Resistance to antibiotics in the normal flora of animals. Vet. Res. 2001, 32, 227–241. [Google Scholar] [CrossRef]
- Stromberg, Z.R.; Van Goor, A.; Redweik, G.A.J.; Brand, M.J.W.; Wannemuehler, M.J.; Mellata, M. Pathogenic and non-pathogenic Escherichia coli colonization and host inflammatory response in a defined microbiota mouse model. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef]
- Animal and Plant Quarantine Agency (APQA). Establishment of Antimicrobial Resistance Surveillance System for Livestock, 2018; Ministry of Agriculture, Food and Rural Affairs: Gimcheon, Korea; Sejong, Korea, 2019.

| No. | Strain | Serovar | Source | Antimicrobial Resistance | Phenotype | ESC Resistance Gene | Conjugation | Plasmid Type |
|---|---|---|---|---|---|---|---|---|
| 1 | A17-KCI-MNK-002-2S | S. Albany | Feces | AMC/AMP/FOX/TAZ/XNL/SXT/FIS/CHL/NAL/TET | AmpC | CMY-2 | a | I1 |
| 2 | A17-KCI-OP-004-4S | S. spp | Feces | AMP/TAZ/XNL/FEP/FIS/NAL/STR/TET | ESBL | CTX-M-15 | NA | ND |
| 3 | A17-KCI-CRBR-001-5 | S. Enteritidis | Carcass | AMP/TAZ/XNL/FEP/FIS/NAL/STR/GEN/TET | ESBL | CTX-M-15 | b | FIIS |
| 4 | A17-KCI-HMR-001-4 | S. Enteritidis | Carcass | AMP/XNL/SXT/FIS/CHL/CIP/NAL/STR/GEN/TET/COL | ESBL | TEM-1, CTX-M-15 | NA | ND |
| 5 | A17-KCI-HMR-002-1 | S. Enteritidis | Carcass | AMP/TAZ/XNL/FEP/NAL/GEN/TET | ESBL | CTX-M-15 | NA | ND |
| 6 | A17-KCI-HMR-002-2 | S. Enteritidis | Carcass | AMP/TAZ/XNL/FEP/NAL/GEN/TET | ESBL | CTX-M-15 | NA | ND |
| 7 | A17-KCI-HMR-002-3 | S. Enteritidis | Carcass | AMP/TAZ/XNL/FEP/NAL/GEN/TET | ESBL | CTX-M-15 | b | FIIS |
| 8 | A17-KCI-HMR-002-4 | S. Enteritidis | Carcass | AMP/TAZ/XNL/FEP/NAL/GEN/TET | ESBL | CTX-M-15 | b | FIIS |
| 9 | A17-KCI-HMR-002-5 | S. Enteritidis | Carcass | AMP/TAZ/XNL/FEP/NAL/GEN/TET | ESBL | CTX-M-15 | NA | ND |
| 10 | A18-KCI-HMR-001-1S | S. Virchow | Feces | AMP/TAZ/XNL/FEP/FIS/NAL/STR/TET | ESBL | CTX-M-15 | b | Nontypeable |
| 11 | A18-KCI-HMR-001-3S | S. Virchow | Feces | AMP/TAZ/XNL/FEP/SXT/FIS/CHL/NAL/STR/TET | ESBL | CTX-M-15 | NA | ND |
| 12 | A18-KCI-HMR-001-4S | S. Virchow | Feces | AMP/TAZ/XNL/FEP/FIS/NAL/STR/TET | ESBL | CTX-M-15 | b | Nontypeable |
| 13 | A18-KCI-HMR-002-2S | S. Virchow | Feces | AMP/TAZ/XNL/FEP/FIS/NAL/STR/TET | ESBL | CTX-M-15 | b | Nontypeable |
| 14 | A18-KCI-HMR-002-3S | S. Virchow | Feces | AMP/TAZ/XNL/FEP/SXT/FIS/CHL/NAL/STR/TET | ESBL | CTX-M-15 | NA | ND |
| 15 | A18-KCI-HMR-002-5S | S. Virchow | Feces | AMP/TAZ/XNL/FEP/SXT/FIS/CHL/NAL/STR/TET | ESBL | CTX-M-15 | NA | ND |
| 16 | A18-KCI-OP-003-2S | S. Enteritidis | Feces | AMP/TAZ/XNL/FEP/NAL/TET | ESBL | CTX-M-15 | NA | ND |
| 17 | A18-KCI-OP-003-3S | S. Enteritidis | Feces | AMP/TAZ/XNL/FEP/NAL/TET | ESBL | CTX-M-15 | NA | ND |
| Strain a | Species | MIC (μg/mL) | Phenotype | ESC Resistance Gene | Conjugation Frequency b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOX | XNL | TAZ | FEP | AMP | AMC | TET | ||||||
| ≥32 | ≥8 | ≥16 | ≥16 | ≥32 | ≥32/16 | ≥16 | ||||||
| MNK | Donor | S. Albany | >32 | >8 | >16 | 1 | >64 | >32/16 | 32 | AmpC | CMY-2 | |
| J53 | Recipient | E. coli | 8 | <0.5 | <1 | <0.25 | < 2 | 4/2 | <2 | |||
| Tc.J53 | Transconjugant | >32 | >8 | >16 | <0.25 | >64 | >32/16 | <2 | AmpC | CMY-2 | 1 × 10−3 ± 1 × 10−7 | |
| 13719 (EIEC) | Recipient | E. coli | 8 | <0.5 | <1 | <0.25 | 8 | 4/2 | >128 | |||
| Tc.13719 | Transconjugant | >32 | >8 | >16 | <0.25 | >64 | >32/16 | >128 | AmpC | CMY-2 | 2 × 10−3 ± 1 × 10−3 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, B.-R.; Wei, B.; Cha, S.-Y.; Shang, K.; Zhang, J.-F.; Jang, H.-K.; Kang, M. Characterization of Extended-Spectrum Cephalosporin (ESC) Resistance in Salmonella Isolated from Chicken and Identification of High Frequency Transfer of blaCMY-2 Gene Harboring Plasmid In Vitro and In Vivo. Animals 2021, 11, 1778. https://doi.org/10.3390/ani11061778
Kwon B-R, Wei B, Cha S-Y, Shang K, Zhang J-F, Jang H-K, Kang M. Characterization of Extended-Spectrum Cephalosporin (ESC) Resistance in Salmonella Isolated from Chicken and Identification of High Frequency Transfer of blaCMY-2 Gene Harboring Plasmid In Vitro and In Vivo. Animals. 2021; 11(6):1778. https://doi.org/10.3390/ani11061778
Chicago/Turabian StyleKwon, Bo-Ram, Bai Wei, Se-Yeoun Cha, Ke Shang, Jun-Feng Zhang, Hyung-Kwan Jang, and Min Kang. 2021. "Characterization of Extended-Spectrum Cephalosporin (ESC) Resistance in Salmonella Isolated from Chicken and Identification of High Frequency Transfer of blaCMY-2 Gene Harboring Plasmid In Vitro and In Vivo" Animals 11, no. 6: 1778. https://doi.org/10.3390/ani11061778
APA StyleKwon, B.-R., Wei, B., Cha, S.-Y., Shang, K., Zhang, J.-F., Jang, H.-K., & Kang, M. (2021). Characterization of Extended-Spectrum Cephalosporin (ESC) Resistance in Salmonella Isolated from Chicken and Identification of High Frequency Transfer of blaCMY-2 Gene Harboring Plasmid In Vitro and In Vivo. Animals, 11(6), 1778. https://doi.org/10.3390/ani11061778





