Contribution to Herpesvirus Surveillance in Beaked Whales Stranded in the Canary Islands
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
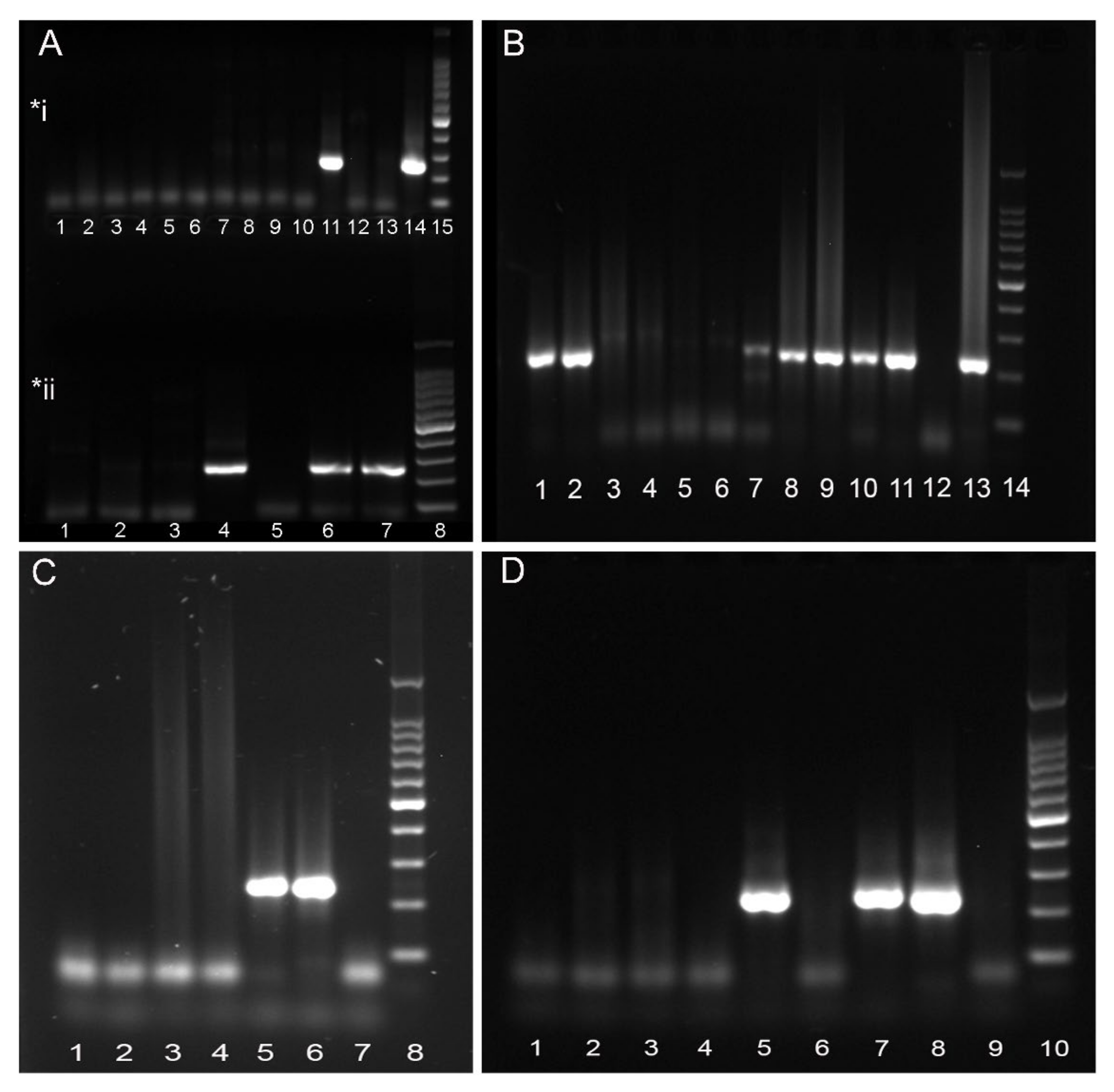
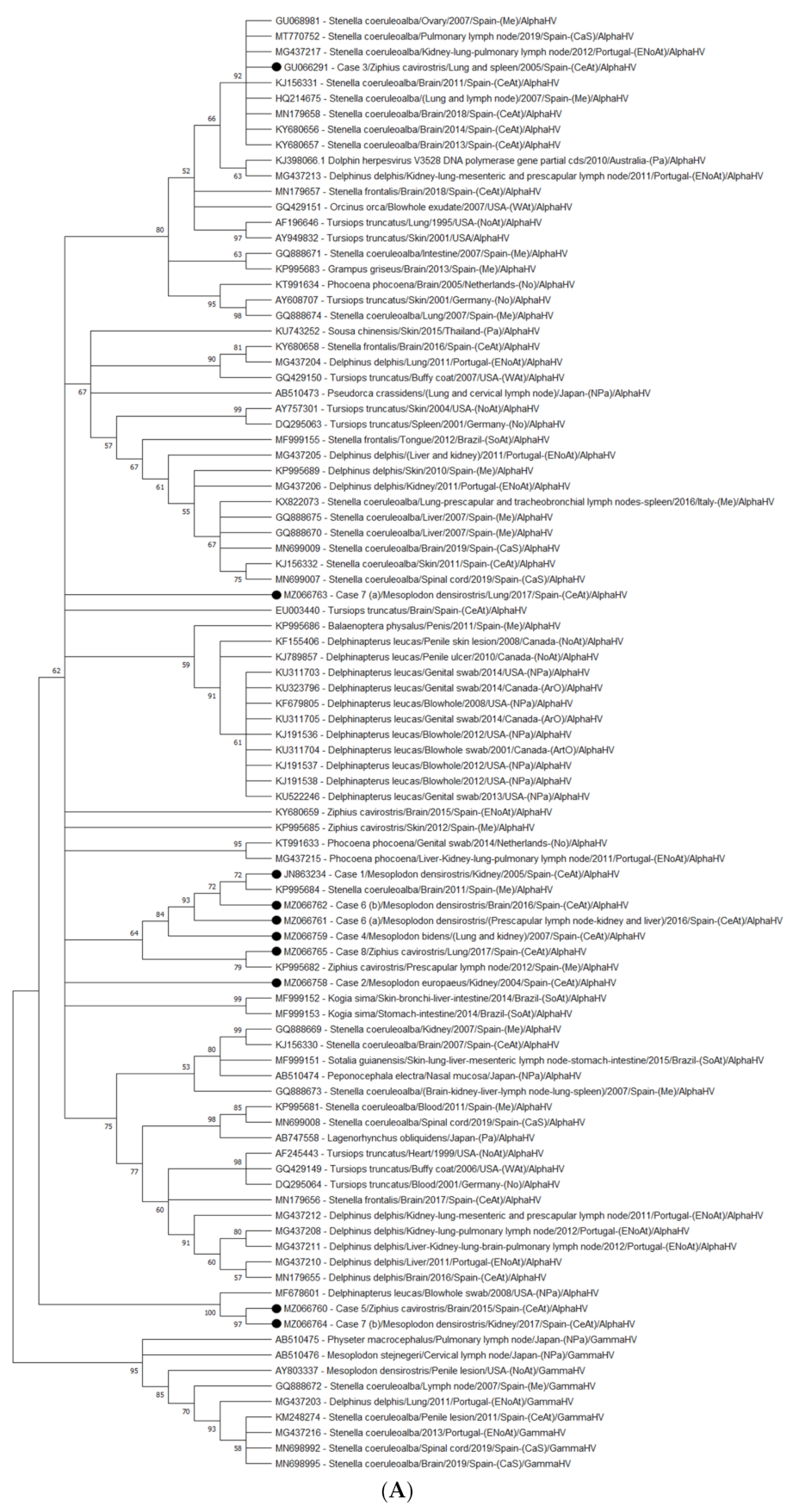
3.1. Molecular Findings
3.1.1. Nucleotide Identity
3.1.2. Amino Acid Identity
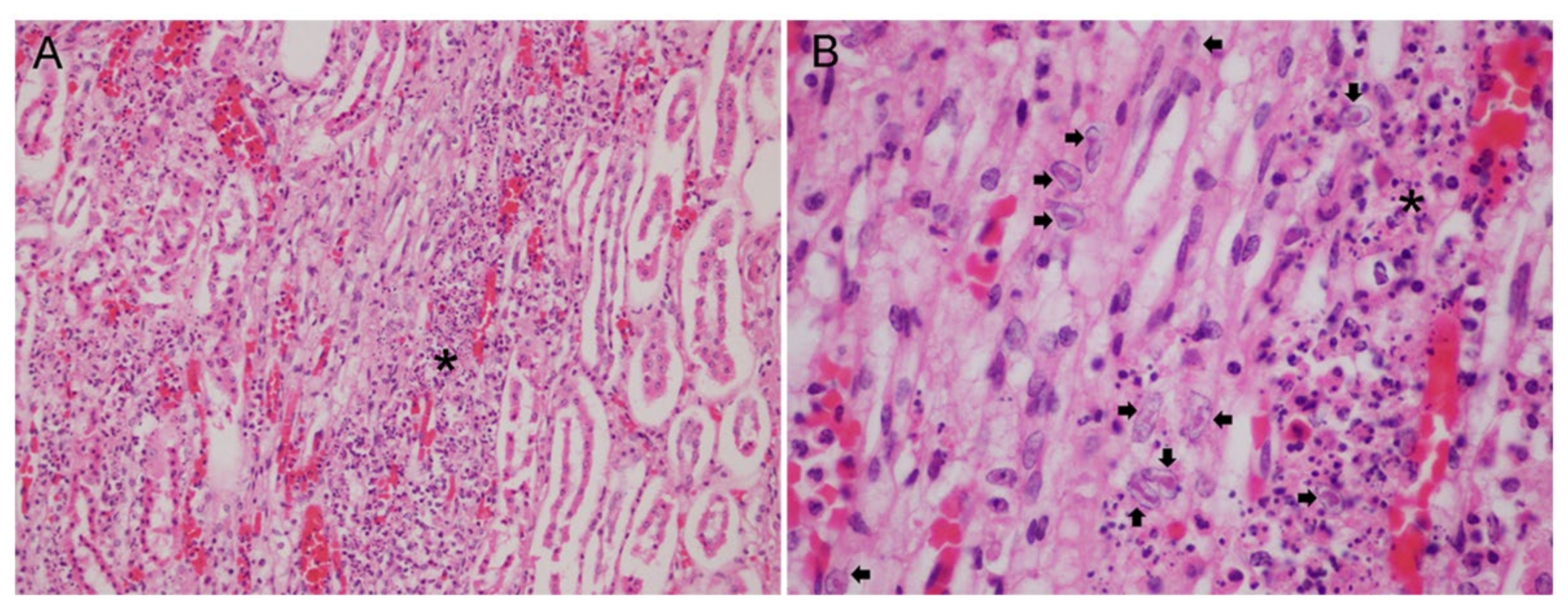
3.2. Gross and Histopathological Findings
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schorr, G.S.; Falcone, E.A.; Moretti, D.J.; Andrews, R.D. First long-term behavioral records from cuvier’s beaked whales (Ziphius cavirostris) reveal record-breaking dives. PLoS ONE 2014, 9, e92633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sierra, E.; Fernandez, A.; Monteros, A.E.D.L.; Arbelo, M.; Díaz-Delgado, J.; Andrada, M.; Herráez, P. Histopathological muscle findings may be essential for a definitive diagnosis of suspected sharp trauma associated with ship strikes in stranded cetaceans. PLoS ONE 2014, 9, e88780. [Google Scholar] [CrossRef] [PubMed]
- Puig-Lozano, R.; Fernandez, A.; Saavedra, P.; Tejedor, M.; Sierra, E.; De La Fuente, J.; Xuriach, A.; Díaz-Delgado, J.; Rivero, M.A.; Andrada, M.; et al. Retrospective study of traumatic intra-interspecific interactions in stranded cetaceans, Canary Islands. Front. Vet.Sci. 2020, 7, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puig-Lozano, R.; Fernández, A.; Sierra, E.; Saavedra, P.; Suárez-Santana, C.M.; De La Fuente, J.; Díaz-Delgado, J.; Godinho, A.; García-Álvarez, N.; Zucca, D.; et al. Retrospective study of fishery interactions in stranded cetaceans, Canary Islands. Front. Vet. Sci. 2020, 7, 567258. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.P.; Jurado, L.F.L. Whales and the military. Nat. Cell Biol. 1991, 351, 448. [Google Scholar] [CrossRef]
- Frantzis, A. Does acoustic testing strand whales? Nat. Cell Biol. 1998, 392, 29. [Google Scholar] [CrossRef]
- Fernández, A.; Edwards, J.F.; Rodríguez, F.; Monteros, A.E.D.L.; Herráez, P.; Castro, P.; Jaber, J.R.; Martín, V.; Arbelo, M. “Gas and Fat Embolic Syndrome” involving a mass stranding of beaked whales (family ziphiidae) exposed to anthropogenic sonar signals. Vet. Pathol. 2005, 42, 446–457. [Google Scholar] [CrossRef]
- Fernandez, A.; Sierra, E.; Martín, V.; Méndez, M.; Sacchinni, S.; De Quirós, Y.B.; Andrada, M.; Rivero, M.; Quesada, O.; Tejedor, M.; et al. Last “Atypical” beaked whales mass stranding in the Canary Islands (July, 2004). J. Mar. Sci. Res. Dev. 2012, 2, 4–6. [Google Scholar] [CrossRef]
- Díaz-Delgado, J.; Fernández, A.; Xuriach, A.; Sierra, E.; De Quirós, Y.B.; Mompeo, B.; Pérez, L.; Andrada, M.; Marigo, J.; Catão-Dias, J.L.; et al. Verminous arteritis due to crassicauda sp. in cuvier’s beaked whales (Ziphius Cavirostris). Vet. Pathol. 2016, 53, 1233–1240. [Google Scholar] [CrossRef] [Green Version]
- Foster, G.; Whatmore, A.; Dagleish, M.P.; Baily, J.; Deaville, R.; Davison, N.J.; Koylass, M.S.; Perrett, L.L.; Stubberfield, E.J.; Reid, R.J.; et al. Isolation of brucella ceti from a long-finned pilot whale (Globicephala melas) and a sowerby’s beaked whale (Mesoploden bidens). J. Wildl. Dis. 2015, 51, 868–871. [Google Scholar] [CrossRef]
- Davison, N.J.; Brownlow, A.; Doeschate, M.T.; Dale, E.-J.; Foster, G.; Muchowski, J.; Perrett, L.L.; Rocchi, M.; Whatmore, A.M.; Dagleish, M.P. Neurobrucellosis due to brucella ceti ST26 in three sowerby’s beaked whales (Mesoplodon bidens). J. Comp. Pathol. 2021, 182, 1–8. [Google Scholar] [CrossRef]
- West, K.L.; Sanchez, S.; Rotstein, D.; Robertson, K.M.; Dennison, S.; Levine, G.; Davis, N.; Schofield, D.; Potter, C.W.; Jensen, B. A Longman’s beaked whale (Indopacetus pacificus) strands in Maui, Hawaii, with first case of morbillivirus in the central Pacific. Mar. Mammal Sci. 2012, 29, 767–776. [Google Scholar] [CrossRef]
- Jacob, J.M.; West, K.L.; Levine, G.; Sanchez, S.; Jensen, B.A. Initial characterization of novel beaked whale morbillivirus in Hawaiian cetaceans. Dis. Aquat. Org. 2016, 117, 215–227. [Google Scholar] [CrossRef] [Green Version]
- Garrigue, C.; Oremus, M.; Dodémont, R.; Bustamante, P.; Kwiatek, O.; Libeau, G.; Lockyer, C.; Vivier, J.C.; Dalebout, M.L. A mass stranding of seven Longman’s beaked whales (Indopacetus pacificus) in New Caledonia, South Pacific. Mar. Mammal Sci. 2016, 32, 884–910. [Google Scholar] [CrossRef]
- Centelleghe, C.; Beffagna, G.; Palmisano, G.; Franzo, G.; Casalone, C.; Pautasso, A.; Giorda, F.; Di Nocera, F.; Iaccarino, D.; Santoro, M.; et al. Dolphin morbillivirus in a cuvier’s beaked whale (Ziphius cavirostris), Italy. Front. Microbiol. 2017, 8, 111. [Google Scholar] [CrossRef]
- Saliki, J.T.; Cooper, E.J.; Rotstein, D.S.; Caseltine, S.L.; Pabst, D.A.; McLellan, W.A.; Govett, P.; Harms, C.; Smolarek, K.A.; Romero, C.H. A Novel Gammaherpesvirus Associated with Genital Lesions in a Blainville’s Beaked Whale (Mesoplodon densirostris). J. Wildl. Dis. 2006, 42, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Arbelo, M.; Sierra, E.; Esperon, F.; Watanabe, T.; Bellière, E.; Monteros, A.E.D.L.; Fernandez, A. Herpesvirus infection with severe lymphoid necrosis affecting a beaked whale stranded in the Canary Islands. Dis. Aquat. Org. 2010, 89, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Arbelo, M.; Bellière, E.N.; Sierra, E.; Sacchinni, S.; Esperón, F.; Andrada, M.; Rivero, M.; Diaz-Delgado, J.; Fernández, A. Herpes virus infection associated with interstitial nephritis in a beaked whale (Mesoplodon densirostris). BMC Vet. Res. 2012, 8, 243. [Google Scholar] [CrossRef] [Green Version]
- International Committee on Taxonomy of Viruses. ICTV Virus Taxonomy: 2019 Release. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 3 September 2020).
- Glaser, R.; Kiecolt-Glaser, J.K. Herpesvirus infections. In Stress-Associated Immune Modulation and Its Implications for Reactivation of Latent Herpesviruses, 1st ed.; Marcel Dekker: New York, NY, USA, 1994; pp. 245–270. ISBN 978-0824788674. [Google Scholar]
- Speck, S.H.; Ganem, D. Viral latency and its regulation: Lessons from the γ-Herpesviruses. Cell Host Microbe 2010, 8, 100–115. [Google Scholar] [CrossRef] [Green Version]
- Penkert, R.R.; Kalejta, R.F. Tegument protein control of latent herpesvirus establishment and animation. Herpesviridae 2011, 2, 3. [Google Scholar] [CrossRef] [Green Version]
- Maclachlan, N.J.; Dubovi, E.J. Herpesvirales. Fenner’s Vet. Virol. 2011, 9, 179–201. [Google Scholar] [CrossRef]
- Bellière, E.N.; Esperon, F.; Arbelo, M.; Muñoz, M.J.; Fernandez, A.; Sánchez-Vizcaíno, J.M. Presence of herpesvirus in striped dolphins stranded during the cetacean morbillivirus epizootic along the Mediterranean Spanish coast in 2007. Arch. Virol. 2010, 155, 1307–1311. [Google Scholar] [CrossRef]
- Blanchard, T.W.; Santiago, N.T.; Lipscomb, T.P.; Garber, R.L.; McFee, W.E.; Knowles, S. Two novel alphaherpesviruses associated with fatal disseminated infections in atlantic bottlenose dolphinS. J. Wildl. Dis. 2001, 37, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Sierra, E.; Sánchez, S.; Saliki, J.; Blas-Machado, U.; Arbelo, M.; Zucca, D.; Fernandez, A. Retrospective study of etiologic agents associated with nonsuppurative meningoencephalitis in stranded cetaceans in the Canary Islands. J. Clin. Microbiol. 2014, 52, 2390–2397. [Google Scholar] [CrossRef] [Green Version]
- Van Elk, C.; Van De Bildt, M.; Van Run, P.; De Jong, A.; Getu, S.; Verjans, G.; Osterhaus, A.; Kuiken, T. Central nervous system disease and genital disease in harbor porpoises (Phocoena phocoena) are associated with different herpesviruses. Vet. Res. 2016, 47, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, S.; Lindstedt, I.J.; Mcaliskey, M.M.; Shirley, A.; Mccullough, S.; Kennedy, S.; Lindstedt, I.J.; Mcaliskey, M.M.; McConnell, S.A.; Mccullough, S.J. Herpesviral Encephalitis in a Harbor Porpoise (Phocoena phocoena). J. Zoo Wildl. Med. 1992, 23, 374–379. [Google Scholar]
- Manire, C.A.; Smolarek, K.A.; Romero, C.H.; Kinsel, M.J.; Clauss, T.M.; Byrd, L. Proliferative dermatitis associated with a novel alphaherpesvirus in an atlantic bottlenose dolphin (Tursiops truncatus). J. Zoo Wildl. Med. 2006, 37, 174–181. [Google Scholar] [CrossRef]
- Benson, K.A.S.; Manire, C.A.; Ewing, R.Y.; Saliki, J.; Townsend, F.I.; Ehlers, B.; Romero, C.H. Identification of novel alpha- and gammaherpesviruses from cutaneous and mucosal lesions of dolphins and whales. J. Virol. Methods 2006, 136, 261–266. [Google Scholar] [CrossRef]
- Sierra, E.; Díaz-Delgado, J.; Arbelo, M.; Andrada, M.; Sacchini, S.; Fernandez, A. Herpesvirus-associated genital lesions in a stranded striped dolphin (Stenella coeruleoalba) in the Canary Islands, Spain. J. Wildl. Dis. 2015, 51, 696–702. [Google Scholar] [CrossRef]
- Van Beurden, S.J.; Ijsseldijk, L.L.; Ordonez, S.R.; Förster, C.; De Vrieze, G.; Gröne, A.; Verheije, M.H.; Kik, M. Identification of a novel gammaherpesvirus associated with (muco)cutaneous lesions in harbour porpoises (Phocoena phocoena). Arch. Virol. 2015, 160, 3115–3120. [Google Scholar] [CrossRef]
- Seade, G.C.C.; Cerqueira, V.D.; Sierra, E.; Chaves, J.F.; Moura, M.A.O.; Montão, D.P.; Riet-Correa, G.; Oliveira, C.A.; Siciliano, S.; Emin-Lima, R.; et al. Herpesviral infection in a Guiana dolphin (Sotalia guianensis) from the northern coast of Brazil. J. Vet. Diagn. Investig. 2017, 29, 877–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vargas-Castro, I.; Crespo-Picazo, J.L.; Rivera-Arroyo, B.; Sánchez, R.; Marco-Cabedo, V.; Jiménez-Martínez, M.Á.; Fayos, M.; Serdio, Á.; García-Párraga, D.; Sánchez-Vizcaíno, J.M. Alpha- and gammaherpesviruses in stranded striped dolphins (Stenella coeruleoalba) from Spain: First molecular detection of gammaherpesvirus infection in central nervous system of odontocetes. BMC Vet. Res. 2020, 16, 1–9. [Google Scholar] [CrossRef]
- Bento, M.; Canha, R.; Eira, C.; Vingada, J.; Nicolau, L.; Ferreira, M.; Domingo, M.; Tavares, L.; Duarte, A. Herpesvirus infection in marine mammals: A retrospective molecular survey of stranded cetaceans in the Portuguese coastline. Infect. Genet. Evol. 2019, 67, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Bellehumeur, C.; Lair, S.; Romero, C.H.; Provost, C.; Nielsen, O.; Gagnon, C.A. Identification of a Novel Herpesvirus Associated with a Penile Proliferative Lesion in a Beluga (Delphinapterus leucas). J. Wildl. Dis. 2015, 51, 244–249. [Google Scholar] [CrossRef] [Green Version]
- Barr, B.; Dunn, J.L.; Daniel, M.D.; Banford, A. Herpes-like viral dermatitis in a beluga whale (Delphinapterus leucas). J. Wildl. Dis. 1989, 25, 608–611. [Google Scholar] [CrossRef] [Green Version]
- Esperón, F.; Fernandez, A.; Sánchez-Vizcaíno, J.M. Herpes simplex-like infection in a bottlenose dolphin stranded in the Canary Islands. Dis. Aquat. Org. 2008, 81, 73–76. [Google Scholar] [CrossRef] [Green Version]
- Lecis, R.; Tocchetti, M.; Rotta, A.; Naitana, S.; Ganges, L.; Pittau, M.; Alberti, A. First gamma herpesvirus detection in a free-living mediterranean bottlenose dolphin. J. Zoo Wildl. Med. 2014, 45, 922–925. [Google Scholar] [CrossRef]
- Miyoshi, K.; Nishida, S.; Sone, E.; Tajima, Y.; Makara, M.; Yoshioka, M.; Nakamura, M.; Yamada, T.K.; Koike, H. Molecular identification of novel alpha- and gammaherpesviruses from cetaceans stranded on Japanese Coasts. Zool. Sci. 2011, 28, 126–133. [Google Scholar] [CrossRef]
- Noguchi, K.; Shimoda, H.; Terada, Y.; Shimojima, M.; Kohyama, K.; Inoshima, Y.; Maeda, K. Isolation of a novel herpesvirus from a Pacific white-sided dolphin. Arch. Virol. 2012, 158, 695–699. [Google Scholar] [CrossRef]
- Melero, M.; Crespo-Picazo, J.L.; Rubio-Guerri, C.; García-Párraga, D.; Sánchez-Vizcaíno, J.M. First molecular determination of herpesvirus from two mysticete species stranded in the Mediterranean Sea. BMC Veter. Res. 2015, 11, 283. [Google Scholar] [CrossRef]
- Van Bressem, M.-F.; Van Waerebeek, K.; Dekegel, D.; Pastoret, P.-P.; Garcia-Godos, A. Herpes-like virus in dusky dolphins, lagenorhynchus obscurus, from coastal Peru. Mar. Mammal Sci. 1994, 10, 354–359. [Google Scholar] [CrossRef]
- Sacristán, C.; Esperon, F.; Ewbank, A.; Díaz-Delgado, J.; Ferreira-Machado, E.; Costa-Silva, S.; Sánchez-Sarmiento, A.M.; Groch, K.R.; Neves, E.; Dutra, G.H.P.; et al. Novel herpesviruses in riverine and marine cetaceans from South America. Acta Trop. 2019, 190, 220–227. [Google Scholar] [CrossRef]
- Soto, S.; González, B.; Willoughby, K.; Maley, M.; Olvera, A.; Kennedy, S.; Marco, A.; Domingo, M. Systemic Herpesvirus and Morbillivirus Co-Infection in a Striped Dolphin (Stenella coeruleoalba). J. Comp. Pathol. 2012, 146, 269–273. [Google Scholar] [CrossRef]
- Rehtanz, M.; Bossart, G.D.; Fair, P.A.; Reif, J.S.; Ghim, S.-J.; Jenson, A.B. Papillomaviruses and herpesviruses: Who is who in genital tumor development of free-ranging Atlantic bottlenose dolphins (Tursiops truncatus)? Vet. Microbiol. 2012, 160, 297–304. [Google Scholar] [CrossRef]
- Cruz, D.; Rodríquez, M.; Kouri, V.; Soto, Y.; Zamora, L.; Rodríguez, D.; Barrera, M.; Rehtanz, M. Concurrent papillomavirus- and herpesvirus-infection in Atlantic bottlenose dolphins (Tursiops truncatus) inhabiting the Cuban coast. Mar. Mammal Sci. 2014, 30, 1564–1572. [Google Scholar] [CrossRef]
- Jsseldijk, L.L.; Brownlow, A.C.; Mazzariol, S. European Best Practice on Cetacean Post mortem Investigation and Tissue Sampling. In Proceedings of the Joint ASCOBANS/ACCOBAMS 2019, 25th Meeting of the Advisory Committee, Stralsund, Germany, 17–19 September 2019. [Google Scholar] [CrossRef]
- Kuiken, T.; García Hartmann, M. Cetacean pathology: Dissection techniques and tissue sampling. In Proceedings of the First ECS Workshop on Cetacean Pathology, ECS Newsletter 17—Special Issue, European Cetacean Society 1991, Leiden, The Netherlands, 13–14 September 1991; pp. 1–39. [Google Scholar]
- Sacristán, C.; Carballo, M.; Muñoz, M.J.; Bellière, E.N.; Neves, E.; Nogal, V.; Esperón, F. Diagnosis of Cetacean morbillivirus: A sensitive one step real time RT fast-PCR method based on SYBR® Green. J. Virol. Methods 2015, 226, 25–30. [Google Scholar] [CrossRef]
- VanDevanter, D.R.; Warrener, P.; Bennett, L.; Schultz, E.R.; Coulter, S.; Garber, R.L.; Rose, T.M. Detection and analysis of diverse herpesviral species by consensus primer PCR. J. Clin. Microbiol. 1996, 34, 1666–1671. [Google Scholar] [CrossRef] [Green Version]
- Groch, K.R.; Taniwaki, S.A.; Favero, C.M.; Brandão, P.E.; Díaz-Delgado, J.; Fernández, A.; Catão-Dias, J.L.; Sierra, E. A novel real-time PCR to detect Cetacean morbillivirus in Atlantic cetaceans. J. Virol. Methods 2020, 285, 113964. [Google Scholar] [CrossRef]
- BLAST: Basic Local Alignment Search Tool. 2021. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 3 May 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hall, B.G. Phylogenetic Trees Made Easy: A How-To Manual, 5th ed.; Oxford University Press: Sunderland, MA, USA, 2017; ISBN 978-0878936069. [Google Scholar]
- Li, R.; Li, Y.; Zheng, H.; Luo, R.; Zhu, H.; Li, Q.; Qian, W.; Ren, Y.; Tian, G.; Li, J.; et al. Building the sequence map of the human pan-genome. Nat. Biotechnol. 2010, 28, 57–63. [Google Scholar] [CrossRef]
- MacLeod, C.D. Beaked Whales, Overview; Elsevier: Amsterdam, The Netherlands, 2018; pp. 80–83. [Google Scholar]
- National Oceanic and Atmospheric Administration. NOAA Fisheries. 2020. Available online: https://www.fisheries.noaa.gov/species/cuviers-beaked-whale (accessed on 17 June 2021).
- National Oceanic and Atmospheric Administration. NOAA Fisheries. 2020. Available online: https://www.fisheries.noaa.gov/species/blainvilles-beaked-whale (accessed on 17 June 2021).
- National Oceanic and Atmospheric Administration. NOAA Fisheries. 2020. Available online: https://www.fisheries.noaa.gov/species/gervais-beaked-whale (accessed on 17 June 2021).
- National Oceanic and Atmospheric Administration. NOAA Fisheries. 2020. Available online: https://www.fisheries.noaa.gov/species/sowerbys-beaked-whale (accessed on 17 June 2021).
- Martín, V.; Tejedor, M.; Pérez-Gil, M.; Dalebout, M.L.; Arbelo, M.; Fernández, A. Records of Gervais’ Beaked Whale Mesoplodon europaeus on the Canary Islands. Eur. Res. Cetaceans 1990, 95, 4. [Google Scholar]
- Martín, V. Short Note: A sowerby’s beaked whale (Mesoplodon bidens) stranded in the Canary Islands: The most southern record in the Eastern North Atlantic. Aquat. Mamm. 2011, 37, 512–519. [Google Scholar] [CrossRef]
- Davison, A.J.; Nielsen, O.; Subramaniam, K.; Jacob, J.M.; Romero, C.H.; Burek-Huntington, K.A.; Waltzek, T.B. Genome Sequence of an Alphaherpesvirus from a Beluga Whale (Delphinapterus leucas). Genome Announc. 2017, 5, e01100-17. [Google Scholar] [CrossRef] [PubMed]
- Ryu, W.-S. Herpesviruses; Elsevier: Amsterdam, The Netherlands, 2017; pp. 125–139. [Google Scholar]
- Maness, H.T.; Nollens, H.H.; Jensen, E.D.; Goldstein, T.; LaMere, S.; Childress, A.; Sykes, J.; Leger, J.S.; Lacave, G.; Latson, F.E.; et al. Phylogenetic analysis of marine mammal herpesviruses. Vet. Microbiol. 2011, 149, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Travers, P.; Walport, M.; Shlomchick, M.; Janeway, C. The Immune System in Health and Disease, 1st ed.; Taylor & Francis: New York, NY, USA, 2001. [Google Scholar]
- Tregoning, J.S.; Schwarze, J. Respiratory viral infections in infants: Causes, clinical symptoms, virology, and immunology. Clin. Microbiol. Rev. 2010, 23, 74–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Species | Locality | AlphaHV | GammaHV |
|---|---|---|---|
| Phocoena phocoena | Sweden, Netherlands, Portugal Northern Hemisphere | [27,28,35] | [27,32] |
| Delphinapterus leucas | Canada | [36,37] | - |
| Tursiops truncatus | Atlantic coast of United States of America (USA) Spain: The Canary Islands Mediterranean Sea | [25,30,38] | [29,30,39] |
| Pseudorca crassidens | Pacific waters | [40] | - |
| Peponocephala electra | Pacific waters | [40] | - |
| Lagenorhynchus obliquidens | Pacific waters | [41] | - |
| Balaenoptera physalus | Mediterranean Sea | [42] | - |
| Stenella coeruleoalba | Portugal Spain: The Canary Islands Cantabrian Sea | [34,35] | [31,34,35] |
| Delphinus delphis | Portugal | [35] | [35] |
| Lagenorhynchus obscurus | South America | [43] | - |
| Stenella frontalis | South America | [44] | - |
| Sotalia guianensis | South America | [44] | [33] |
| Kogia sima | South America | [44] | - |
| Ziphius cavirostris | Spain: The Canary Islands | [17] | - |
| Mesoplodon densirostris | Spain: The Canary Islands Atlantic coast of United States of America (USA) | [18] | [16,30] |
| Grampus griseus | Atlantic coast of United States of America (USA) | - | [30] |
| Physeter macrocephalus | Japanese coast | - | [40] |
| Balaenoptera acutorostrata | Mediterranean Sea | - | [42] |
| Inia boliviensis | South America | - | [44] |
| Mesoplodon stejnegeri | Japanese coast | - | [40] |
| ID CODE | SEX | AGE | SD | SL | SS | DC | TESTED SAMPLES |
|---|---|---|---|---|---|---|---|
| CET 86 | F | A | 27/11/1999 | Tenerife | A | 3 | Skin, lung, liver, kidney |
| CET 103 | M | J | 19/04/2000 | Fuerteventura | D | 3 | Lung, liver, kidney, brain |
| CET 108 | F | A | 10/06/2000 | Tenerife | D | 3 | Skin, skeletal muscle, lung, liver, kidney |
| CET 113 | F | S | 16/07/2000 | Tenerife | D | 3 | Skin, skeletal muscle |
| CET 181 | M | S | 24/09/2002 | Fuerteventura | A | 2 | Skin, skeletal muscle, lung, mediastinal and mesenteric lymph node, liver, kidney, brain, spleen |
| CET 182 | M | S | 24/09/2002 | Fuerteventura | D | 2 | Skin, skeletal muscle, lung, liver, mesenteric lymph node, kidney, brain, spleen |
| CET 183 | M | S | 24/09/2002 | Fuerteventura | D | 2 | Skin, skeletal muscle, liver, mesenteric lymph node, kidney, brain |
| CET 184 | M | S | 24/09/2002 | Fuerteventura | D | 2 | Skin, skeletal muscle, lung, liver, mediastinal and mesenteric lymph node, kidney, brain, spleen, thyroid |
| CET 189 | F | A | 27/09/2002 | Fuerteventura | D | 4 | Skin, lung, liver, kidney |
| CET 236 | F | C | 21/03/2004 | La Graciosa | D | 3 | Skin, skeletal muscle, lung, liver, kidney, brain |
| CET 264 | F | N.D. | 23/07/2004 | Fuerteventura | D | 4 | Liver, skeletal muscle, lung, kidney |
| CET 265 | M | A | 24/07/2004 | Fuerteventura | D | 4 | Skin, skeletal muscle, lung, liver, kidney |
| * CET 294 | F | A | 18/04/2005 | Fuerteventura | D | 4 | Skin, skeletal muscle, lung, liver, spleen |
| CET 304 | F | C | 13/07/2005 | Fuerteventura | D | 2 | Skin, skeletal muscle, lung, liver, kidney |
| CET 322 | M | A | 17/02/2006 | Gran Canaria | D | 4 | Skin, lung, liver, kidney |
| CET 352 | N.D. | J | 06/07/2006 | Tenerife | D | 3 | Lung, kidney, brain, spleen |
| CET 471 | F | S | 06/11/2008 | Fuerteventura | D | 2 | Lung, kidney, brain, spleen |
| CET 503 | F | A | 21/09/2009 | Gran Canaria | D | 4 | Lung, kidney |
| CET 576 | F | A | 16/05/2011 | Lanzarote | D | 2 | Lung, kidney, brain, spleen |
| CET 579 | M | S | 13/06/2011 | Tenerife | D | 4 | Lung, mesenteric lymph node, kidney, brain, spleen |
| CET 591 | F | A | 01/11/2011 | Tenerife | D | 4 | Lung, prescapular lymph node, kidney, brain, spleen |
| CET 593 | M | A | 18/11/2011 | Gran Canaria | D | 4 | Skin, lung, prescapular lymph node, liver, kidney |
| CET 620 | M | A | 20/05/2012 | Gran Canaria | D | 4 | Skin, lung, liver, kidney, spleen |
| CET 624 | F | A | 13/07/2012 | La Graciosa | D | 3 | Skin, lung, liver, mesenteric lymph node, kidney, brain |
| CET 645 | M | J | 09/02/2013 | Lanzarote | D | 4 | Skin, liver, brain |
| CET 680 | F | N | 02/07/2013 | Gran Canaria | D | 4 | Lung, intestine, mesenteric lymph node, kidney, brain, spleen |
| CET 688 | F | A | 18/11/2013 | Gran Canaria | D | 4 | Brain |
| CET 712 | F | S | 28/04/2014 | Fuerteventura | D | 4 | Prescapular lymph node, spleen |
| CET 719 | F | A | 06/06/2014 | Lanzarote | D | 3 | Lung, mesenteric lymph node, kidney, spleen |
| CET 720 | N.D. | S | 10/06/2014 | Fuerteventura | D | 4 | Lung, mesenteric lymph node, kidney, brain |
| CET 770 | M | S | 28/07/2015 | Tenerife | D | 3 | Lung, intestine, mesenteric lymph node, brain, spleen |
| CET 771 | F | A | 05/08/2015 | Tenerife | D | 2 | Lung, intestine, mesenteric lymph node, kidney, brain, spleen |
| CET 818 | M | S | 16/08/2016 | Gran Canaria | D | 4 | Lung, intestine, mesenteric lymph node, kidney, brain, spleen |
| CET 833 | N.D. | N.D. | 13/02/2017 | Tenerife | D | 4 | Lung, mesenteric lymph node, kidney, brain |
| CET 855 | M | A | 22/05/2017 | Gran Canaria | D | 3 | Lung, intestine, mesenteric lymph node, kidney, brain, spleen |
| ID CODE | SPECIES | SEX | AGE | SD | SL | SS | DC | TESTED SAMPLES |
|---|---|---|---|---|---|---|---|---|
| CET 134 | M. europaeus | F | C | 28/06/2001 | Gran Canaria | A | 1 | Skin, lung, liver, kidney, brain |
| CET 180 | M. densirostris | F | A | 24/09/2002 | Fuerteventura | A | 2 | Skin |
| CET 185 | M. europaeus | F | A | 24/09/2002 | Fuerteventura | D | 2 | Skin, skeletal muscle, lung, liver, mesenteric lymph node, kidney, brain, spleen |
| CET 213 | M. densirostris | F | A | 28/06/2003 | Gran Canaria | A | 1 | Skin, skeletal muscle, lung, liver, kidney, brain |
| * CET 243 | M. densirostris | M | A | 18/04/2004 | Tenerife | A | 1 | Skin, skeletal muscle, lung, liver, kidney |
| CET 259 | M. europaeus | F | J | 21/06/2004 | Fuerteventura | D | 2 | Skin, skeletal muscle, lung, liver, kidney, brain |
| CET 333 | M. europaeus | F | S | 28/03/2006 | El Hierro | A | 2 | Skin, skeletal muscle, lung, thymus, liver, mesenteric lymph node, kidney, brain, spleen |
| CET 334 | M. europaeus | F | S | 28/03/2006 | El Hierro | A | 2 | Skin, skeletal muscle, lung, liver, kidney, brain, spleen |
| CET 338 | M. europaeus | F | J | 06/04/2006 | Gran Canaria | D | 2 | Skin, skeletal muscle, lung, liver, blood, mesenteric lymph node, kidney, brain, spleen |
| CET 354 | M. europaeus | M | C | 28/07/2006 | Tenerife | D | 4 | Skin, lung, liver, kidney, spleen |
| CET 379 | M. bidens | M | A | 16/04/2007 | Lanzarote | D | 2 | Skin, lung, liver, kidney, brain, spleen |
| CET 510 | M. europaeus | M | A | 14/12/2009 | Lanzarote | D | 2 | Skin, lung, liver, mesenteric lymph node, kidney, brain |
| CET 547 | M. europaeus | M | A | 29/08/2010 | Fuerteventura | D | 4 | Skin, lung, liver, mesenteric lymph node, kidney, brain |
| CET 631 | M. europaeus | M | A | 21/10/2012 | Fuerteventura | D | 4 | Skin, lung, penis, palate, esophagus, brain |
| CET 636 | M. mirus | M | S | 30/11/2012 | El Hierro | D | 2 | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen |
| CET 695 | M. densirostris | F | A | 12/07/2014 | Lanzarote | D | 4 | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen |
| CET 711 | M. densirostris | M | S | 03/04/2014 | El Hierro | D | 5 | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen |
| CET 824 | M. densirostris | F | A | 11/11/2016 | Fuerteventura | D | 2 | Skin, prescapular lymph node, liver, kidney, brain, spleen |
| CET 827 | M. bidens | F | A | 07/12/2016 | La Gomera | A | 4 | Skin, lung, liver, mesenteric lymph node, intestine, brain, spleen |
| CET 852 | M. densirostris | F | A | 05/05/2017 | Fuerteventura | D | 2 | Skin, lung, liver, mesenteric lymph node, kidney, brain, spleen |
| CASE n° | ID CODE | SPECIES | SEX | AGE | SD | SL | SS | DC | HV-POSITIVE SAMPLES | HV-OBTAINED SEQUENCES |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | * CET 243 | M. de | M | A | 18/04/2002 | T | A | 1 | Lung, kidney | (Arbelo et al., 2012) |
| 2 | CET 259 | M. eu | F | A | 21/06/2004 | F | D | 2 | Kidney | MZ066758 |
| 3 | * CET 294 | Z. ca | F | A | 18/04/2005 | F | D | 4 | Lung, spleen | (Arbelo et al., 2010) |
| 4 | CET 379 | M. bi | M | A | 16/04/2007 | L | D | 2 | Lung, kidney | MZ066759 |
| 5 | CET 771 | Z. ca | F | A | 06/08/2015 | T | D | 2 | Brain | MZ066760 |
| 6 | CET 824 | M. de | F | A | 11/11/2016 | F | D | 2 | (a) Liver, prescapular lymph node, kidney; (b) brain | (a) MZ066761, (b) MZ066762 |
| 7 | CET 852 | M. de | F | A | 05/05/2017 | F | D | 2 | (a) Lung; (b) kidney | (a) MZ066763, (b) MZ066764 |
| 8 | CET 855 | Z. ca | M | A | 22/05/2017 | GC | D | 3 | Lung | MZ066765 |
| * Case 3 | Case 5 | Case 8 | |
|---|---|---|---|
| CET | 294 | 771 | 855 |
| GenBank Acc. No. | GU066291 | MZ066760 | MZ066765 |
| Samples | Lung and spleen | Brain | Lung |
| Nucleotide identity | 100% MG437217 (S. co); KY680657 (S.co); KY680656 (S. co); KJ156331 (S. co) | 98.48% KP995682 (Z. ca) | 98.06% KP995682 (Z. ca) |
| Aminoacid identity | 100% AUZ97325 (S. co) AUZ97326 (S. co) AHN91834 (S. co) | 98.48% ALP00298 (Z. ca) | 98.53% ALP00298 (Z. ca) |
| * Case 1 | Case 2 | Case 4 | Case 6 | Case 7 | |
|---|---|---|---|---|---|
| CET | 243 | 259 | 379 | 824 | 852 |
| GenBank Acc. No. | JN863234 | MZ066758 | MZ066759 | (a) MZ066761 (b) MZ066762 | (a) MZ066763 (b) MZ066764 |
| Species | M. densirostris | M. europaeus | M. bidens | M. densirostris | M. densirostris |
| Samples | Lung and kidney | Kidney | Lung and kidney | (a) Liver, prescapular lymph node and kidney (b) Brain | (a) Lung (b) Kidney |
| Nucleotide identity | 95.65% KP995684 (S.co) | 78.35% KP995682 (Z. ca) | 89.32% JN863234 (M. de) | (a) 93.59% JN863234 (M. de) (b) 97.04% KP995684 (S. co) | (a) 78.3% KF155406 (D. le) (b) 97.46% KP995684 (S.co) |
| Amino acid identity | 79.13% ANG08598 (D. le) | 82.81% ALP00298 (Z. ca) | 95.59% ALP00300 (S.co) | (a) 97.06% ALP00300 (S.co) (b) 97.06% ALP00300 (S.co) | (a) 72.73% ALP00292 (Z. ca) (b) 96.92% ALP00300 (S.co) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felipe-Jiménez, I.; Fernández, A.; Andrada, M.; Arbelo, M.; Segura-Göthlin, S.; Colom-Rivero, A.; Sierra, E. Contribution to Herpesvirus Surveillance in Beaked Whales Stranded in the Canary Islands. Animals 2021, 11, 1923. https://doi.org/10.3390/ani11071923
Felipe-Jiménez I, Fernández A, Andrada M, Arbelo M, Segura-Göthlin S, Colom-Rivero A, Sierra E. Contribution to Herpesvirus Surveillance in Beaked Whales Stranded in the Canary Islands. Animals. 2021; 11(7):1923. https://doi.org/10.3390/ani11071923
Chicago/Turabian StyleFelipe-Jiménez, Idaira, Antonio Fernández, Marisa Andrada, Manuel Arbelo, Simone Segura-Göthlin, Ana Colom-Rivero, and Eva Sierra. 2021. "Contribution to Herpesvirus Surveillance in Beaked Whales Stranded in the Canary Islands" Animals 11, no. 7: 1923. https://doi.org/10.3390/ani11071923
APA StyleFelipe-Jiménez, I., Fernández, A., Andrada, M., Arbelo, M., Segura-Göthlin, S., Colom-Rivero, A., & Sierra, E. (2021). Contribution to Herpesvirus Surveillance in Beaked Whales Stranded in the Canary Islands. Animals, 11(7), 1923. https://doi.org/10.3390/ani11071923






