Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profiles in Testes of Calves and Sexually Mature Wandong Bulls (Bos taurus)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. Histological Study of Testicular Tissues
2.3. RNA Isolation and Integrity Assessment
2.4. cDNA Library Construction for lncRNA-mRNAs Sequencing Analysis
2.5. Transcriptome Assembly
2.6. LncRNAs and mRNAs Identification and Coding Potential
2.7. Expression Pattern of Differentially Expressed lncRNAs and mRNAs
2.8. GO and KEGG Pathways Enrichments
2.9. Cis and Trans-Regulated Genes Prediction
2.10. Validation of DE lncRNAs and mRNAs Gene via RT-qPCR
2.11. Statistical Analysis
3. Results
3.1. The Morphological Study of Testicular Tissues
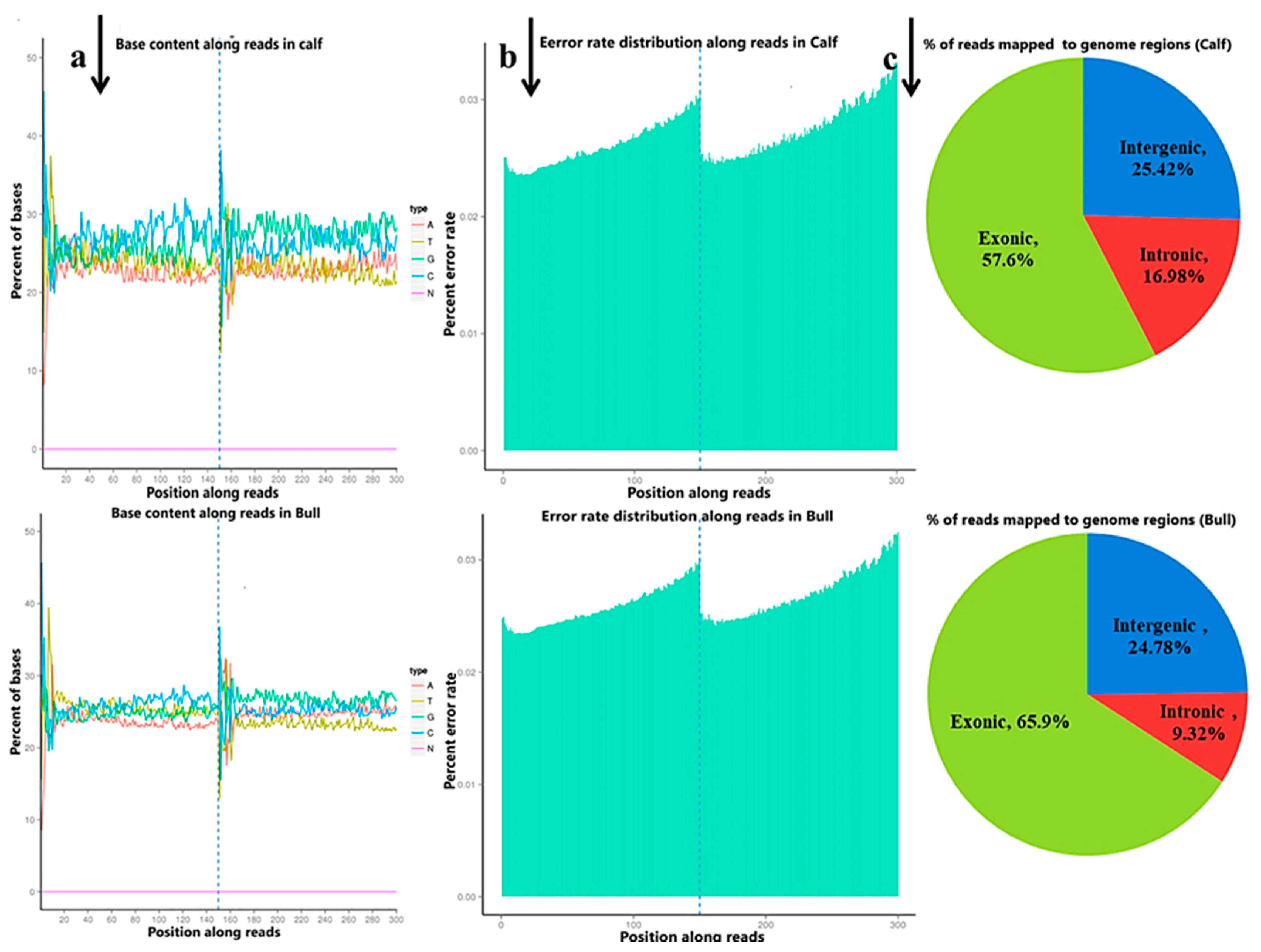
3.2. Outline of lncRNAs and mRNAs Sequencing in Calf and Bull Testes
3.3. Identification and Characterizations of lncRNAs and mRNAs
3.4. Characteristics Comparison between lncRNAs and mRNAs
3.5. Differentially Expressed Gene-Level lncRNAs and mRNAs in Testicular Tissues
3.6. Functional Annotation and Enrich KEGG Pathways Analysis of DE mRNAs
3.7. LncRNAs Co-Location and Co-Expression Regulated Target Genes
3.8. Validation of Differentially Expressed LncRNAs and mRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuwissen, T.H.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- König, S.; Simianer, H.; Willam, A. Economic evaluation of genomic breeding programs. J. Dairy Sci. 2009, 92, 382–391. [Google Scholar] [CrossRef] [PubMed]
- Hecht, N.B. Molecular mechanisms of male germ cell differentiation. Bioessays 1998, 20, 555–561. [Google Scholar] [CrossRef]
- Luk, A.; Chan, W.-Y.; Rennert, O.M.; Lee, T.-L. Long noncoding RNAs in spermatogenesis: Insights from recent high-throughput transcriptome studies. Reproduction 2014, 147, R131–R141. [Google Scholar] [CrossRef] [Green Version]
- Chocu, S.; Calvel, P.; Rolland, A.D.; Pineau, C. Spermatogenesis in mammals: Proteomic insights. Syst. Biol. Reprod. Med. 2012, 58, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, C.; Guo, F. Integrated analysis of mRNAs and long noncoding RNAs in the semen from Holstein bulls with high and low sperm motility. Sci. Rep. 2019, 9, 2092. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Li, S.; Lai, Z.; Zhou, Z.; Wu, F.; Huang, Y.; Lan, X.; Lei, C.; Chen, H.; Dang, R. Analysis of Long Non-Coding RNA and mRNA Expression Profiling in Immature and Mature Bovine (Bos taurus) Testes. Front. Genet. 2019, 10, 646. [Google Scholar] [CrossRef] [Green Version]
- Weng, B.; Ran, M.; Chen, B.; He, C.; Dong, L.; Peng, F. Genome-wide analysis of long non-coding RNAs and their role in postnatal porcine testis development. Genomics 2017, 109, 446–456. [Google Scholar] [CrossRef]
- Yang, H.; Wang, F.; Li, F.; Ren, C.; Pang, J.; Wan, Y.; Wang, Z.; Feng, X.; Zhang, Y. Comprehensive analysis of long noncoding RNA and mRNA expression patterns in sheep testicular maturation. Biol. Reprod. 2018, 99, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Rinn, J.; Guttman, M. RNA and dynamic nuclear organization. Science 2014, 345, 1240–1241. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhu, H.; Song, W.; Li, M.; Liu, C.; Li, N.; Tang, F.; Mu, H.; Liao, M.; Li, X. Identification of conservative microRNAs in Saanen dairy goat testis through deep sequencing. Reprod. Domest. Anim. 2014, 49, 32–40. [Google Scholar] [CrossRef]
- Guan, Y.; Liang, G.; Hawken, P.A.; Malecki, I.A.; Cozens, G.; Vercoe, P.E.; Martin, G.B. Roles of small RNAs in the effects of nutrition on apoptosis and spermatogenesis in the adult testis. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, C.; Romero, Y.; Warnefors, M.; Bilican, A.; Borel, C.; Smith, L.B.; Kotaja, N.; Kaessmann, H.; Nef, S. Germ cell-specific targeting of DICER or DGCR8 reveals a novel role for endo-siRNAs in the progression of mammalian spermatogenesis and male fertility. PLoS ONE 2014, 9, e107023. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L.; Zhao, J.C. Functional analysis of long noncoding RNAs in development and disease. Syst. Biol. RNA Bind. Proteins 2014, 129–158. [Google Scholar] [CrossRef]
- Xu, J.; Bai, J.; Zhang, X.; Lv, Y.; Gong, Y.; Liu, L.; Zhao, H.; Yu, F.; Ping, Y.; Zhang, G. A comprehensive overview of lncRNA annotation resources. Brief. Bioinform. 2017, 18, 236–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dianatpour, A.; Ghafouri-Fard, S. Long non coding RNA expression intersecting cancer and spermatogenesis: A systematic review. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 2601. [Google Scholar] [PubMed]
- Sun, J.; Lin, Y.; Wu, J. Long non-coding RNA expression profiling of mouse testis during postnatal development. PLoS ONE 2013, 8, e75750. [Google Scholar] [CrossRef]
- Wen, K.; Yang, L.; Xiong, T.; Di, C.; Ma, D.; Wu, M.; Xue, Z.; Zhang, X.; Long, L.; Zhang, W. Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res. 2016, 26, 1233–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, M.; Chen, B.; Li, Z.; Wu, M.; Liu, X.; He, C.; Zhang, S.; Li, Z. Systematic identification of long noncoding RNAs in immature and mature porcine testes. Biol. Reprod. 2016, 94, 71–79. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, Y.; Li, Y.; Bai, H.; Xue, F.; Xu, S.; Xu, H.; Shi, L.; Yang, N.; Chen, J. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility. Sci. Rep. 2017, 7, 9055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Yang, H.; Han, L.; Li, F.; Zhang, T.; Pang, J.; Feng, X.; Ren, C.; Mao, S.; Wang, F. Long noncoding RNA expression profile changes associated with dietary energy in the sheep testis during sexual maturation. Sci. Rep. 2017, 7, 5180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arun, G.; Akhade, V.S.; Donakonda, S.; Rao, M.R.S. mrhl RNA, a long noncoding RNA, negatively regulates Wnt signaling through its protein partner Ddx5/p68 in mouse spermatogonial cells. Mol Cell. Biol. 2012, 32, 3140–3152. [Google Scholar] [CrossRef] [Green Version]
- Anguera, M.C.; Ma, W.; Clift, D.; Namekawa, S.; Kelleher, R.J., III; Lee, J.T. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 2011, 7, e1002248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agbor, V.A.; Tao, S.; Lei, N.; Heckert, L.L. A Wt1-Dmrt1 transgene restores DMRT1 to Sertoli cells of Dmrt1−/− testes: A novel model of DMRT1-deficient germ cells. Biol. Reprod. 2013, 88. [Google Scholar] [CrossRef] [Green Version]
- Lunstra, D.-D.; Echternkamp, S. Puberty in beef bulls: Acrosome morphology and semen quality in bulls of different breeds. J. Anim. Sci. 1982, 55, 638–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Hematoxylin and eosin staining of tissue and cell sections. Cold Spring Harb. Protoc. 2008, 2008, pdb-prot4986. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Guttman, M.; Garber, M.; Levin, J.Z.; Donaghey, J.; Robinson, J.; Adiconis, X.; Fan, L.; Koziol, M.J.; Gnirke, A.; Nusbaum, C. Ab initio reconstruction of cell type–specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010, 28, 503–510. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [Green Version]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.-E.; Song, H.K. Identification of tissue-enriched novel transcripts and novel exons in mice. BMC Genomics 2014, 15, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, A.; Zhang, J.; Zhou, Z.; Wang, L.; Liu, Y.; Liu, Y. ALDB: A domestic-animal long noncoding RNA database. PLoS ONE 2015, 10, e0124003. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.D.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E.L. The Pfam protein families database. Nucleic Acids Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Flexible analysis of transcriptome assemblies with Ballgown. Biorxiv 2014. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Reuveni, E.; Getselter, D.; Oron, O.; Elliott, E. Differential contribution of cis and trans gene transcription regulatory mechanisms in amygdala and prefrontal cortex and modulation by social stress. Sci. Rep. 2018, 8, 6339. [Google Scholar] [CrossRef] [Green Version]
- Ghanbarian, A.T.; Hurst, L.D. Neighboring genes show correlated evolution in gene expression. Mol. Biol. Evol. 2015, 32, 1748–1766. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocol. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Veneziano, D.; Nigita, G.; Ferro, A. Computational approaches for the analysis of ncRNA through deep sequencing techniques. Front. Bioeng. Biotechnol. 2015, 3, 77. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Gao, L.; Xu, E.Y. LncRNA, a new component of expanding RNA-protein regulatory network important for animal sperm development. Semin. Cell Dev. Biol. 2016, 59, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Wu, J.; Schuster, A.S.; Hennig, G.W.; Yan, W. Expression profiling reveals developmentally regulated lncRNA repertoire in the mouse male germline. Biol. Reprod. 2013, 89, 107-1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoneda, R.; Satoh, Y.; Yoshida, I.; Kawamura, S.; Kotani, T.; Kimura, A.P. A genomic region transcribed into a long noncoding RNA interacts with the Prss42/Tessp-2 promoter in spermatocytes during mouse spermatogenesis, and its flanking sequences can function as enhancers. Mol. Reprod. Dev. 2016, 83, 541–557. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.H.; Kwon, J.T.; Kim, J.; Jeong, J.; Kim, J.; Lee, S.; Cho, C. Profiling of testis-specific long noncoding RNAs in mice. BMC Genomics 2018, 19, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, X.; He, Y.; Chen, S.-Y.; Wang, J.; Hu, S.; Lai, S.-J. Genome-wide identification and characterisation of long non-coding RNAs in two Chinese cattle breeds. Ital. J. Anim. Sci. 2020, 19, 383–391. [Google Scholar] [CrossRef]
- Liu, Y.; Li, M.; Bo, X.; Li, T.; Ma, L.; Zhai, T.; Huang, T. Systematic analysis of Long non-coding RNAs and mRNAs in the ovaries of Duroc pigs during different follicular stages using RNA sequencing. Int. J. Mol. Sci. 2018, 19, 1722. [Google Scholar] [CrossRef] [Green Version]
- Zeng, B.; Chen, T.; Xie, M.-Y.; Luo, J.-Y.; He, J.-J.; Xi, Q.-Y.; Sun, J.-J.; Zhang, Y.-L. Exploration of long noncoding RNA in bovine milk exosomes and their stability during digestion in vitro. J. Dairy Sci. 2019, 102, 6726–6737. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Li, C.; Liu, S.; Zhou, C.; Yin, H.; Song, J.; Zhang, Q.; Zhang, S. Genome wide identification of novel long non-coding RNAs and their potential associations with milk proteins in Chinese Holstein cows. Front. Genet. 2018, 9, 281. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Do, D.N.; Dudemaine, P.-L.; Fomenky, B.E.; Bissonnette, N. Integration of lncRNA and mRNA transcriptome analyses reveals genes and pathways potentially involved in calf intestinal growth and development during the early weeks of life. Genes 2018, 9, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.; Luo, Y.; Liu, M.; Huang, J.; Xu, D. Histological and transcriptome analyses of testes from Duroc and Meishan boars. Sci. Rep. 2016, 6, 20758. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hong, Y.; Yuan, C.; Fu, Z.; Shi, Y.; Zhang, M.; Shen, L.; Han, Y.; Zhu, C.; Li, H. Microarray analysis of gene expression profiles of Schistosoma japonicum derived from less-susceptible host water buffalo and susceptible host goat. PLoS ONE 2013, 8, e70367. [Google Scholar] [CrossRef] [Green Version]
- Young, J.C.; Wakitani, S.; Loveland, K.L. TGF-β superfamily signaling in testis formation and early male germline development. Semin. Cell Dev. Biol. 2015, 45, 94–103. [Google Scholar] [CrossRef]
- Li, T.; Zhang, H.; Wang, X.; Yin, D.; Chen, N.; Kang, L.; Zhao, X.; Ma, Y. Cloning, molecular characterization and expression patterns of DMRTC2 implicated in germ cell development of male Tibetan sheep. Int. J. Mol. Sci. 2020, 21, 2448. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Wang, J.; Wang, K.; Luo, Y.; Tang, Q.; Liu, X.; Fang, M. Integrated Analysis of miRNA-mRNA Network Reveals Different Regulatory Patterns in the Endometrium of Meishan and Duroc Sows during Mid-Late Gestation. Animals 2020, 10, 420. [Google Scholar] [CrossRef] [Green Version]
- Nixon, B.; Bromfield, E.G.; Dun, M.D.; Redgrove, K.A.; McLaughlin, E.A.; Aitken, R.J. The role of the molecular chaperone heat shock protein A2 (HSPA2) in regulating human sperm-egg recognition. Asian J. Androl. 2015, 17, 568. [Google Scholar] [CrossRef]
- Scieglinska, D.; Krawczyk, Z. Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans. Cell Stress Chaperones 2015, 20, 221–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Wu, M.; Fan, Y.; Li, S.; Lai, Z.; Huang, Y.; Lan, X.; Lei, C.; Chen, H.; Dang, R. Identification and characterization of circular RNAs in Qinchuan cattle testis. R. Soc. Open Sci. 2018, 5, 180413. [Google Scholar] [CrossRef] [Green Version]
- Li, R.-K.; Tan, J.-L.; Chen, L.-T.; Feng, J.-S.; Liang, W.-X.; Guo, X.-J.; Liu, P.; Chen, Z.; Sha, J.-H.; Wang, Y.-F. Iqcg is essential for sperm flagellum formation in mice. PLoS ONE 2014, 9, e98053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Houmard, B.; Small, C.; Yang, L.; Naluai-Cecchini, T.; Cheng, E.; Hassold, T.; Griswold, M. Global gene expression in the human fetal testis and ovary. Biol. Reprod. 2009, 81, 438–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sambroni, E.; Rolland, A.D.; Lareyre, J.-J.; Le Gac, F. Fsh and Lh have common and distinct effects on gene expression in rainbow trout testis. J. Mol. Endocrinol. 2013, 50, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Wang, F. Genes related with male gonadal morphogenesis in mammals. Zhonghua Nan Ke Xue Natl. J. Androl. 2008, 14, 356–359. [Google Scholar]
- Ponting, C.P.; Oliver, P.L.; Reik, W. Evolution and functions of long noncoding RNAs. Cell 2009, 136, 629–641. [Google Scholar] [CrossRef] [Green Version]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
- Wu, Q.; Song, R.; Yan, W. SPATA3 and SPATA6 Interact with KLHL10 and Participate in Spermatogenesis; Oxford University Press: Oxford, UK, 2010. [Google Scholar]






| DE-lnc-RNA/ID | Chromosome/Location | Lnc-RNA-Status | Regulation | DE cis-Regulated Genes/ID | Gene-Symbol | Chromosome/Location | Enrich Pathways |
|---|---|---|---|---|---|---|---|
| TCONS_00223595 | 25: 40984697– 41226938 | Novel-lncRNA | Up | ENSBTAG00000002474 | MAD1L1 | 25: 40984696– 41222673 | Cell Cycle |
| TCONS_00218376 | 24: 47452790– 47459159 | Novel-lncRNA | Down | ENSBTAG00000039916 | SMAD2 | 24: 47500431– 47586992 | Cell Cycle |
| TCONS_00285708 | 4: 49484422– 49563305 | Novel-lncRNA | Down | ENSBTAG00000006732 | NRCAM | 4: 49248702– 49563305 | Cell Adhesion Molecules (CAMs) |
| TCONS_00265960 | 3: 3819912– 3824649 | Novel-lncRNA | Down | ENSBTAG00000012025 | LMX1A | 3: 3653710– 3828022 | Cell Adhesion Molecules (CAMs) |
| TCONS_00263842 | 3: 1.02 × 108– 1.02 × 108 | Novel-lncRNA | Up | ENSBTAG00000000253 | PTPRF | 3: 102414018–102498351 | Adherens Junction |
| TCONS_00092191 | 15: 30147243– 30156216 | Novel-lncRNA | Down | ENSBTAG00000015939 | NECTIN1 | 15: 30211169– 30282836 | Adherens Junction |
| TCONS_00226910 | 25: 35113818– 35117275 | Novel-lncRNA | Up | ENSBTAG00000026273 | MYL10 | 25: 35177976– 35190739 | Tight Junction |
| TCONS_00003412 | 1: 53213665– 53221154 | Novel-lncRNA | Up | ENSBTAG00000018399 | MYH15 | 1: 53114698– 53273181 | Tight Junction |
| TCONS_00314718 | 6: 98071217– 98093465 | Novel-lncRNA | Down | ENSBTAG00000005745 | HPSE | 6: 98071126– 98118656 | Metabolic Pathway |
| TCONS_00305722 | 5: 76121498– 76122025 | Novel-lncRNA | Down | ENSBTAG00000030632 | ALG10 | 5: 76104065– 76113301 | Metabolic Pathway |
| TCONS_00106232 | 16: 56589861– 56596165 | Novel-lncRNA | Down | ENSBTAG00000011706 | TNR | 16: 56545152– 56764975 | ECM-receptor Interaction |
| TCONS_00340198 | 7: 75125739– 75137127 | Novel-lncRNA | Up | ENSBTAG00000014773 | HMMR | 7: 75137017– 75184925 | ECM-receptor Interaction |
| TCONS_00282996 | 4: 11670810– 11680838 | Novel-lncRNA | Down | ENSBTAG00000013472 | COL1A2 | 4: 11776162– 11823181 | Focal Adhesion |
| ENSBTAT00000080812 | 5: 32333604– 32349222 | Annotated-lncRNA | Up | ENSBTAG00000013155 | COL2A1 | 5: 32283180– 32313172 | Focal Adhesion |
| TCONS_00098825 | 15: 37889979– 37909497 | Novel-lncRNA | Up | ENSBTAG00000005218 | PDE3B | 15: 37935445– 38104280 | cAMP Signaling Pathway |
| TCONS_00267075 | 3: 16683665– 16685238 | Novel-lncRNA | Up | ENSBTAG00000006234 | NPR1 | 3: 16683665– 16700685 | cAMP Signaling Pathway |
| TCONS_00118360 | 17: 53611398– 53625902 | Novel-lncRNA | Down | ENSBTAG00000004457 | ORAI1 | 17: 53610136– 53626039 | Calcium Signaling Pathway |
| TCONS_00038312 | 11: 22365812– 22406197 | Novel-lncRNA | Down | ENSBTAG00000013861 | SLC8A1 | 11: 22408368– 22787993 | Calcium Signaling Pathway |
| TCONS_00310078 | 6: 11726041– 11732204 | Novel-lncRNA | Down | ENSBTAG00000014463 | CAMK2D | 6: 11800357– 12189732 | GnRH Signaling Pathway |
| TCONS_00321828 | 6: 1.01 × 108– 1.01 × 108 | Novel-lncRNA | Down | ENSBTAG00000020048 | MAPK10 | 6: 100907869–101279063 | GnRH Signaling Pathway |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Khan, I.M.; Yin, H.; Zhou, X.; Rizwan, M.; Zhuang, J.; Zhang, Y. Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profiles in Testes of Calves and Sexually Mature Wandong Bulls (Bos taurus). Animals 2021, 11, 2006. https://doi.org/10.3390/ani11072006
Liu H, Khan IM, Yin H, Zhou X, Rizwan M, Zhuang J, Zhang Y. Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profiles in Testes of Calves and Sexually Mature Wandong Bulls (Bos taurus). Animals. 2021; 11(7):2006. https://doi.org/10.3390/ani11072006
Chicago/Turabian StyleLiu, Hongyu, Ibrar Muhammad Khan, Huiqun Yin, Xinqi Zhou, Muhammad Rizwan, Jingyi Zhuang, and Yunhai Zhang. 2021. "Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profiles in Testes of Calves and Sexually Mature Wandong Bulls (Bos taurus)" Animals 11, no. 7: 2006. https://doi.org/10.3390/ani11072006
APA StyleLiu, H., Khan, I. M., Yin, H., Zhou, X., Rizwan, M., Zhuang, J., & Zhang, Y. (2021). Integrated Analysis of Long Non-Coding RNA and mRNA Expression Profiles in Testes of Calves and Sexually Mature Wandong Bulls (Bos taurus). Animals, 11(7), 2006. https://doi.org/10.3390/ani11072006






