Dysregulation of PI3K/Akt/PTEN Pathway in Canine Mammary Tumor
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sample Collection and Histopathological Evaluation
2.3. DNA Extraction
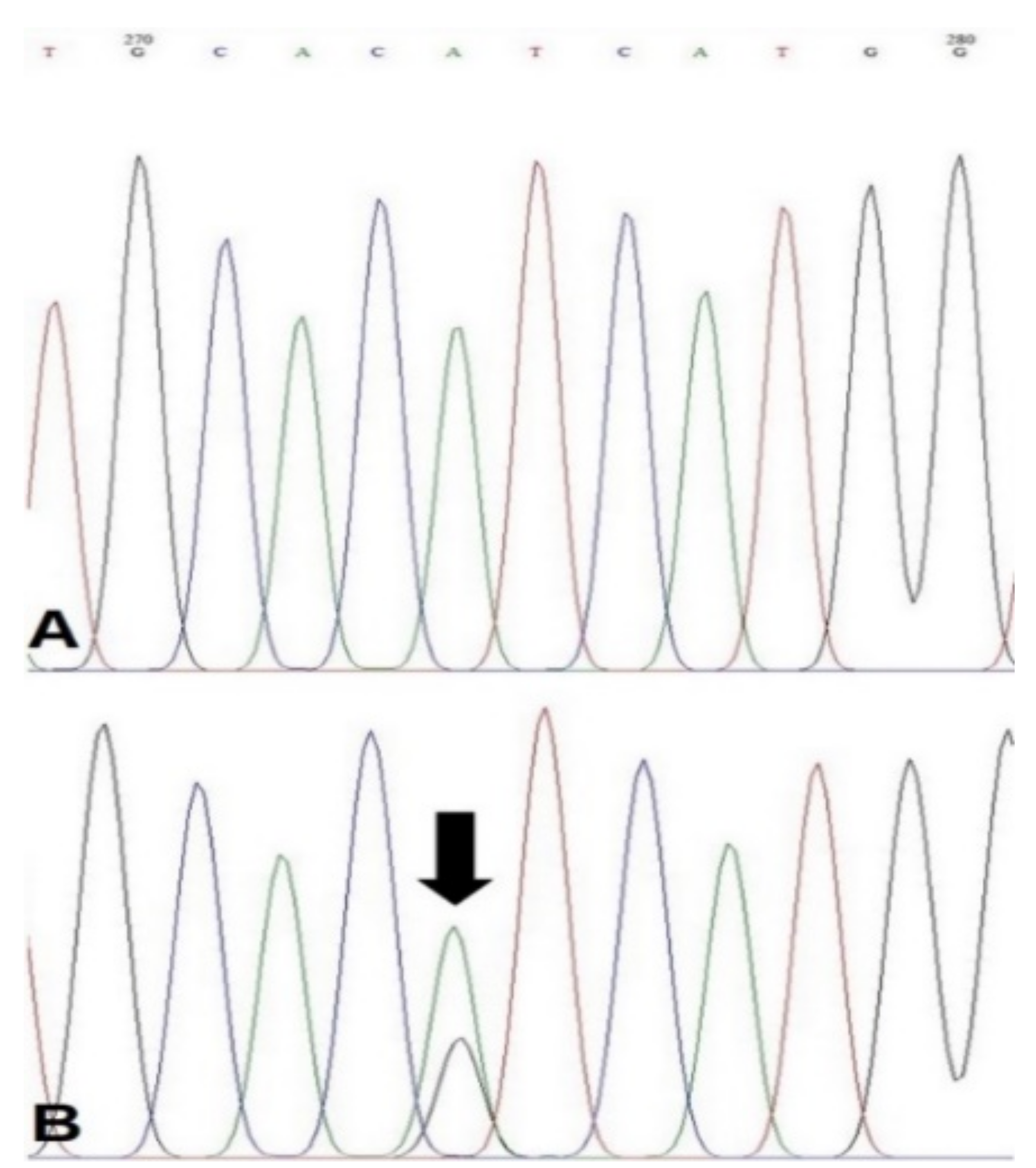
2.4. PCR and Sequence Analysis
2.5. Immunohistochemistry (IHC)
2.5.1. Controls for IHC
2.5.2. IHC
2.6. RNA In Situ Hybridization (RISH)
2.7. Evaluation of IHC and RISH
2.8. Statistical Analysis
3. Results
3.1. Histopathological Features of Samples
3.2. Frequency of PIK3CA H1047R Mutation and Relevance to Marker Expressions
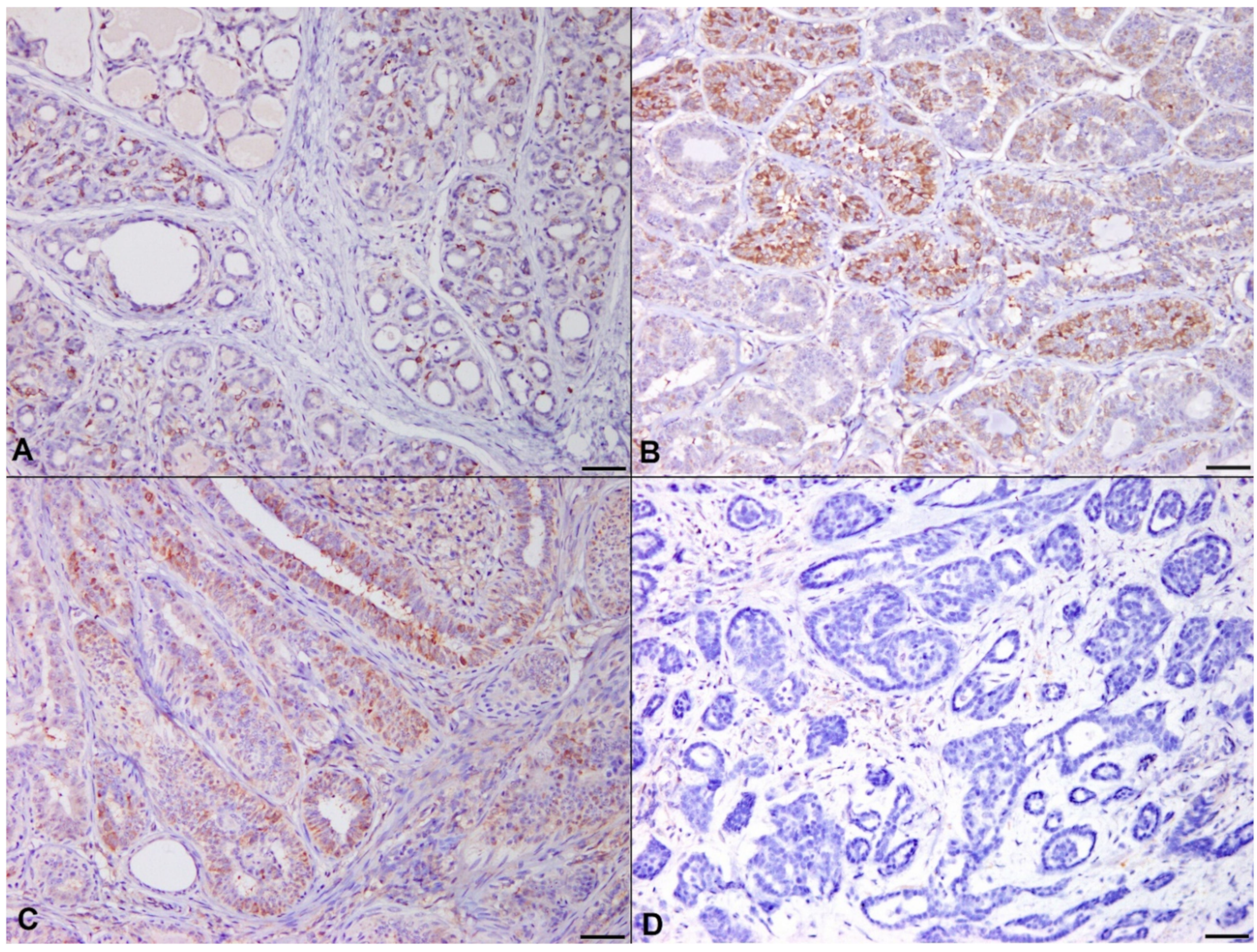
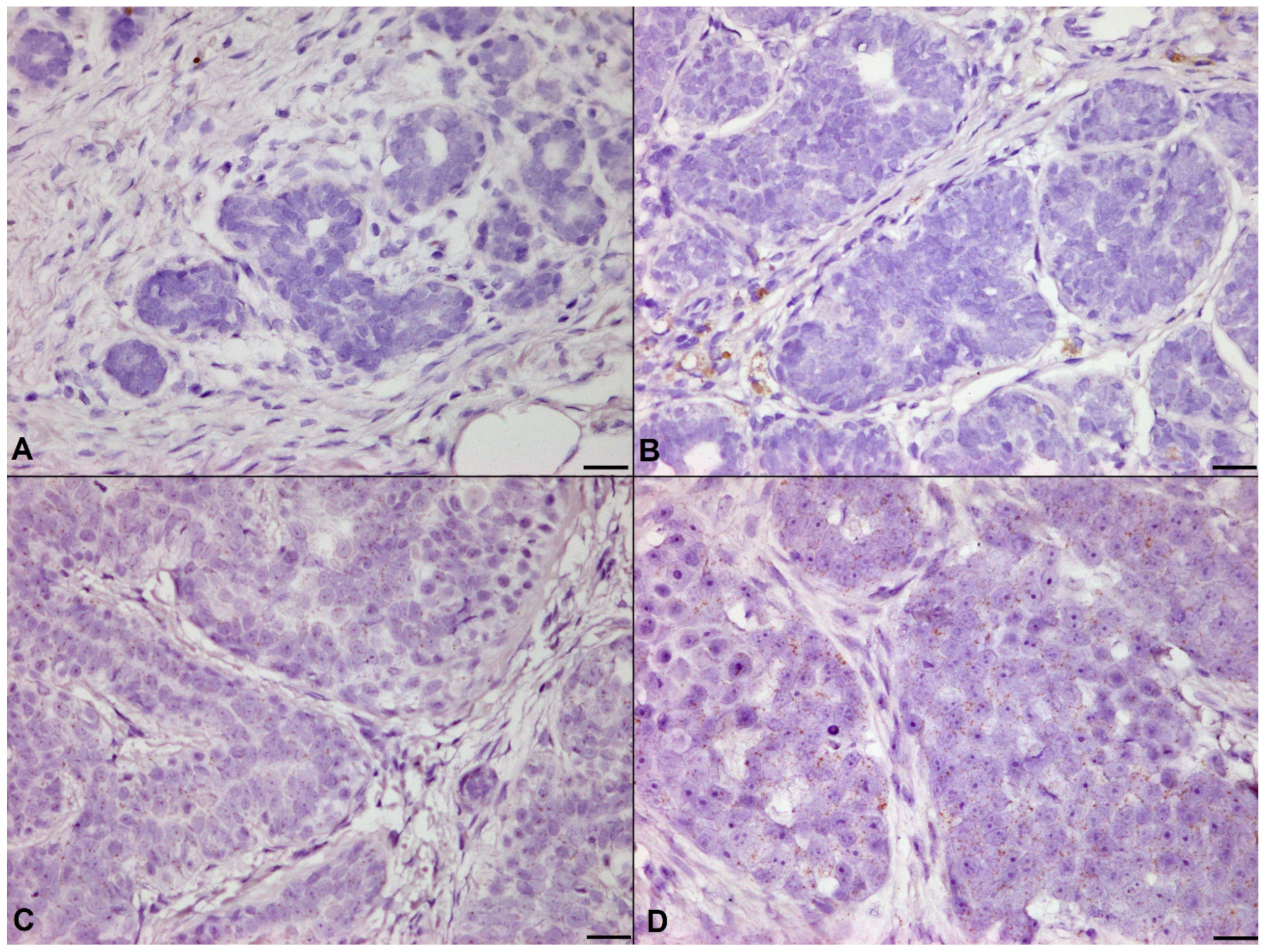
3.3. Controls for IHC
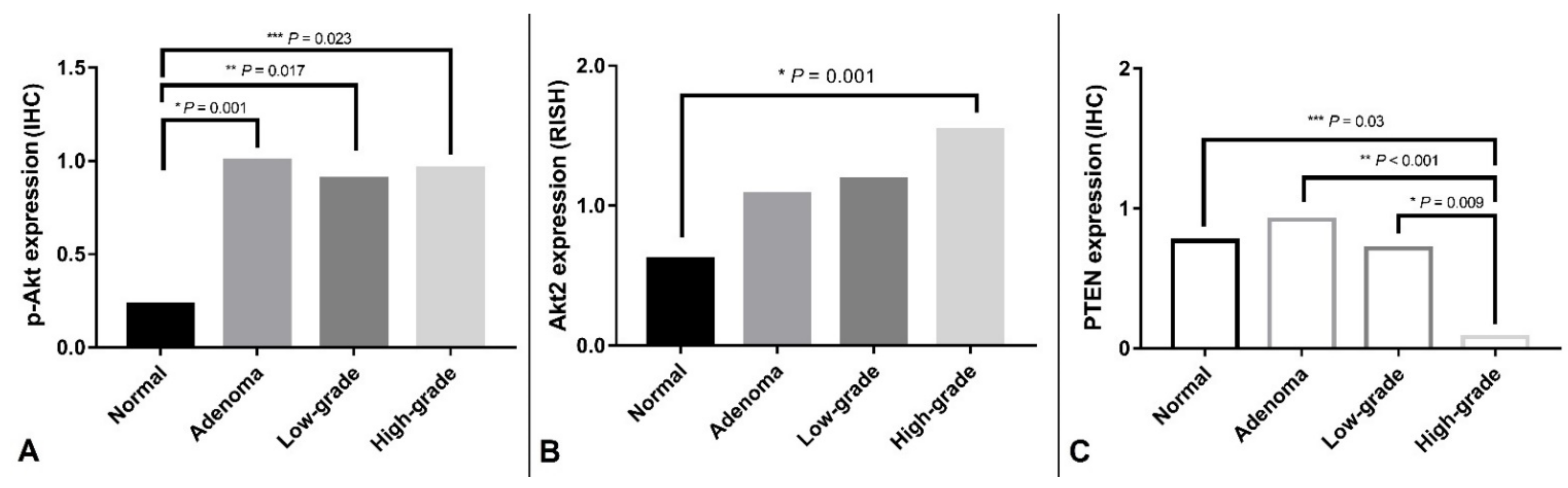
3.4. Correlation of Histological Grade with PIK3CA H1047R Mutation and IHC/RISH Marker Expressions
3.5. Histopathological/Clinical Features and PIK3CA Mutation/Marker Expressions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arafeh, R.; Samuels, Y. PIK3CA in Cancer: The Past 30 Years. Semin. Cancer Biol. 2019, 59, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.; Kiger, A.A. Classes of Phosphoinositide 3-Kinases at a Glance. J. Cell Sci. 2014, 127, 923–928. [Google Scholar] [CrossRef] [Green Version]
- Martini, M.; De Santis, M.C.; Braccini, L.; Gulluni, F.; Hirsch, E. PI3K/AKT Signaling Pathway and Cancer: An Updated Review. Ann. Med. 2014, 46, 372–383. [Google Scholar] [CrossRef]
- Paez, J.; Sellers, W.R. PI3K/PTEN/AKT Pathway. A Critical Mediator of Oncogenic Signaling. Cancer Treat. Res. 2003, 115, 145–167. [Google Scholar]
- Jerde, T.J. Phosphatase and Tensin Homologue: Novel Regulation by Developmental Signaling. J. Signal. Transduct. 2015, 2015, 282567. [Google Scholar] [CrossRef]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fruman, D.A.; Rommel, C. PI3K and Cancer: Lessons, Challenges and Opportunities. Nat. Rev. Drug Discov. 2014, 13, 140–156. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.; Lee, J.T.; Navolanic, P.M.; Steelman, L.S.; Shelton, J.G.; Blalock, W.L.; Franklin, R.A.; McCubrey, J.A. Involvement of PI3K/Akt Pathway in Cell Cycle Progression, Apoptosis, and Neoplastic Transformation: A Target for Cancer Chemotherapy. Leukemia 2003, 17, 590–603. [Google Scholar] [CrossRef] [Green Version]
- Cain, R.J.; Ridley, A.J. Phosphoinositide 3-Kinases in Cell Migration. Biol. Cell 2009, 101, 13–29. [Google Scholar] [CrossRef]
- Carnero, A.; Paramio, J.M. The PTEN/PI3K/AKT Pathway in Vivo, Cancer Mouse Models. Front. Oncol. 2014, 4, 252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, I.G.; Russell, S.E.; Choong, D.Y.H.; Montgomery, K.G.; Ciavarella, M.L.; Hooi, C.S.F.; Cristiano, B.E.; Pearson, R.B.; Phillips, W.A. Mutation of the PIK3CA Gene in Ovarian and Breast Cancer. Cancer Res. 2004, 64, 7678–7681. [Google Scholar] [CrossRef] [Green Version]
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High Frequency of Mutations of the PIK3CA Gene in Human Cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartmann, C.; Bartels, G.; Gehlhaar, C.; Holtkamp, N.; von Deimling, A. PIK3CA Mutations in Glioblastoma Multiforme. Acta Neuropathol. 2005, 109, 639–642. [Google Scholar] [CrossRef]
- Polom, K.; Marrelli, D.; Roviello, G.; Pascale, V.; Voglino, C.; Vindigni, C.; Generali, D.; Roviello, F. PIK3CA Mutation in Gastric Cancer and the Role of Microsatellite Instability Status in Mutations of Exons 9 and 20 of the PIK3CA Gene. Adv. Clin. Exp. Med. 2018, 27, 963–969. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Li, J.; Li, J.; Che, G. Clinical Significance of PIK3CA Gene in Non-Small-Cell Lung Cancer: A Systematic Review and Meta-Analysis. BioMed Res. Int. 2020, 2020, 3608241. [Google Scholar] [CrossRef]
- Rosty, C.; Young, J.P.; Walsh, M.D.; Clendenning, M.; Sanderson, K.; Walters, R.J.; Parry, S.; Jenkins, M.A.; Win, A.K.; Southey, M.C.; et al. PIK3CA Activating Mutation in Colorectal Carcinoma: Associations with Molecular Features and Survival. PLoS ONE 2013, 8, e65479. [Google Scholar] [CrossRef] [Green Version]
- Arsenic, R.; Lehmann, A.; Budczies, J.; Koch, I.; Prinzler, J.; Kleine-Tebbe, A.; Schewe, C.; Loibl, S.; Dietel, M.; Denkert, C. Analysis of PIK3CA Mutations in Breast Cancer Subtypes. Appl. Immunohistochem. Mol. Morphol. 2014, 22, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Kalinsky, K.; Jacks, L.M.; Heguy, A.; Patil, S.; Drobnjak, M.; Bhanot, U.K.; Hedvat, C.V.; Traina, T.A.; Solit, D.; Gerald, W.; et al. PIK3CA Mutation Associates with Improved Outcome in Breast Cancer. Clin. Cancer Res. 2009, 15, 5049–5059. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and Spectrum of PIK3CA Somatic Mutations in Breast Cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.-L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An Integrative Genomic and Proteomic Analysis of PIK3CA, PTEN, and AKT Mutations in Breast Cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H. PIK3CA Mutations Matter for Cancer in Dogs. Res. Vet. Sci. 2020, 133, 39–41. [Google Scholar] [CrossRef]
- Adams, J.R.; Xu, K.; Liu, J.C.; Agamez, N.M.R.; Loch, A.J.; Wong, R.G.; Wang, W.; Wright, K.L.; Lane, T.F.; Zacksenhaus, E.; et al. Cooperation between Pik3ca and P53 Mutations in Mouse Mammary Tumor Formation. Cancer Res. 2011, 71, 2706–2717. [Google Scholar] [CrossRef] [Green Version]
- Koren, S.; Reavie, L.; Couto, J.P.; De Silva, D.; Stadler, M.B.; Roloff, T.; Britschgi, A.; Eichlisberger, T.; Kohler, H.; Aina, O.; et al. PIK3CA(H1047R) Induces Multipotency and Multi-Lineage Mammary Tumours. Nature 2015, 525, 114–118. [Google Scholar] [CrossRef]
- Meyer, D.S.; Brinkhaus, H.; Müller, U.; Müller, M.; Cardiff, R.D.; Bentires-Alj, M. Luminal Expression of PIK3CA Mutant H1047R in the Mammary Gland Induces Heterogeneous Tumors. Cancer Res. 2011, 71, 4344–4351. [Google Scholar] [CrossRef] [Green Version]
- Dong, L.; Meng, F.; Wu, L.; Mitchell, A.V.; Block, C.J.; Zhang, B.; Craig, D.B.; Jang, H.; Chen, W.; Yang, Q.; et al. Cooperative Oncogenic Effect and Cell Signaling Crosstalk of Co-occurring HER2 and Mutant PIK3CA in Mammary Epithelial Cells. Int. J. Oncol. 2017, 51, 1320–1330. [Google Scholar] [CrossRef] [Green Version]
- Alsaihati, B.A.; Ho, K.-L.; Watson, J.; Feng, Y.; Wang, T.; Zhao, S. Canine Tumor Mutation Rate Is Positively Correlated with TP53 Mutation across Cancer Types and Breeds. bioRxiv 2020, 205286. [Google Scholar] [CrossRef]
- Kim, T.-M.; Yang, I.S.; Seung, B.-J.; Lee, S.; Kim, D.; Ha, Y.-J.; Seo, M.-K.; Kim, K.-K.; Kim, H.S.; Cheong, J.-H.; et al. Cross-Species Oncogenic Signatures of Breast Cancer in Canine Mammary Tumors. Nat. Commun. 2020, 11, 3616. [Google Scholar] [CrossRef]
- Lee, K.-H.; Hwang, H.-J.; Noh, H.J.; Shin, T.-J.; Cho, J.-Y. Somatic Mutation of PIK3CA (H1047R) Is a Common Driver Mutation Hotspot in Canine Mammary Tumors as Well as Human Breast Cancers. Cancers 2019, 11, 2006. [Google Scholar] [CrossRef] [Green Version]
- Asproni, P.; Millanta, F.; Ressel, L.; Podestà, F.; Parisi, F.; Vannozzi, I.; Poli, A. An Immunohistochemical Study of the PTEN/AKT Pathway Involvement in Canine and Feline Mammary Tumors. Animals 2021, 11, 365. [Google Scholar] [CrossRef]
- Ressel, L.; Millanta, F.; Caleri, E.; Innocenti, V.M.; Poli, A. Reduced PTEN Protein Expression and Its Prognostic Implications in Canine and Feline Mammary Tumors. Vet. Pathol. 2009, 46, 860–868. [Google Scholar] [CrossRef]
- Goldschmidt, M.; Peña, L.; Rasotto, R.; Zappulli, V. Classification and Grading of Canine Mammary Tumors. Vet. Pathol. 2011, 48, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, S.M.; Baskin, D.G.; Frevert, C.W.; Stahl, W.L.; Rosa-Molinar, E. Controls for Immunohistochemistry: The Histochemical Society’s Standards of Practice for Validation of Immunohistochemical Assays. J. Histochem. Cytochem. 2014, 62, 693–697. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- The Human Protein Atlas. Available online: https://www.proteinatlas.org/ (accessed on 25 May 2021).
- Murai, A.; Abou Asa, S.; Kodama, A.; Sakai, H.; Hirata, A.; Yanai, T. Immunohistochemical Analysis of the Akt/MTOR/4E-BP1 Signalling Pathway in Canine Haemangiomas and Haemangiosarcomas. J. Comp. Pathol. 2012, 147, 430–440. [Google Scholar] [CrossRef]
- Hinz, N.; Jücker, M. Distinct Functions of AKT Isoforms in Breast Cancer: A Comprehensive Review. Cell Commun. Signal. 2019, 17, 154. [Google Scholar] [CrossRef] [Green Version]
- Jang, S.-H.; Kim, K.-J.; Oh, M.-H.; Lee, J.-H.; Lee, H.J.; Cho, H.D.; Han, S.W.; Son, M.W.; Lee, M.S. Clinicopathological Significance of Elevated PIK3CA Expression in Gastric Cancer. J. Gastric Cancer 2016, 16, 85–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuboki, Y.; Schatz, C.A.; Koechert, K.; Schubert, S.; Feng, J.; Wittemer-Rump, S.; Ziegelbauer, K.; Krahn, T.; Nagatsuma, A.K.; Ochiai, A. In Situ Analysis of FGFR2 MRNA and Comparison with FGFR2 Gene Copy Number by Dual-Color in Situ Hybridization in a Large Cohort of Gastric Cancer Patients. Gastric Cancer 2018, 21, 401–412. [Google Scholar] [CrossRef] [Green Version]
- Mizoguchi, M.; Betensky, R.A.; Batchelor, T.T.; Bernay, D.C.; Louis, D.N.; Nutt, C.L. Activation of STAT3, MAPK, and AKT in Malignant Astrocytic Gliomas: Correlation with EGFR Status, Tumor Grade, and Survival. J. Neuropathol. Exp. Neurol. 2006, 65, 1181–1188. [Google Scholar] [CrossRef] [Green Version]
- Scrima, M.; De Marco, C.; Fabiani, F.; Franco, R.; Pirozzi, G.; Rocco, G.; Ravo, M.; Weisz, A.; Zoppoli, P.; Ceccarelli, M.; et al. Signaling Networks Associated with AKT Activation in Non-Small Cell Lung Cancer (NSCLC): New Insights on the Role of Phosphatydil-Inositol-3 Kinase. PLoS ONE 2012, 7, e30427. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Tenorio, G.; Alkhori, L.; Olsson, B.; Waltersson, M.A.; Nordenskjöld, B.; Rutqvist, L.E.; Skoog, L.; Stål, O. PIK3CA Mutations and PTEN Loss Correlate with Similar Prognostic Factors and Are Not Mutually Exclusive in Breast Cancer. Clin. Cancer Res. 2007, 13, 3577–3584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maruyama, N.; Miyoshi, Y.; Taguchi, T.; Tamaki, Y.; Monden, M.; Noguchi, S. Clinicopathologic Analysis of Breast Cancers with PIK3CA Mutations in Japanese Women. Clin. Cancer Res. 2007, 13, 408. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, K.; Mahajan, N.P. PI3K-Independent AKT Activation in Cancers: A Treasure Trove for Novel Therapeutics. J. Cell Physiol. 2012, 227, 3178–3184. [Google Scholar] [CrossRef] [Green Version]
- Arsenic, R.; Treue, D.; Lehmann, A.; Hummel, M.; Dietel, M.; Denkert, C.; Budczies, J. Comparison of Targeted Next-Generation Sequencing and Sanger Sequencing for the Detection of PIK3CA Mutations in Breast Cancer. BMC Clin. Pathol. 2015, 15, 20. [Google Scholar] [CrossRef] [Green Version]
- Qiu, C.; Lin, D.; Wang, J.; Wang, L. Expression and Significance of PTEN in Canine Mammary Gland Tumours. Res. Vet. Sci. 2008, 85, 383–388. [Google Scholar] [CrossRef]
- Milella, M.; Falcone, I.; Conciatori, F.; Cesta Incani, U.; Del Curatolo, A.; Inzerilli, N.; Nuzzo, C.M.A.; Vaccaro, V.; Vari, S.; Cognetti, F.; et al. PTEN: Multiple Functions in Human Malignant Tumors. Front. Oncol. 2015, 5, 24. [Google Scholar] [CrossRef] [Green Version]
- Rasotto, R.; Berlato, D.; Goldschmidt, M.H.; Zappulli, V. Prognostic Significance of Canine Mammary Tumor Histologic Subtypes: An Observational Cohort Study of 229 Cases. Vet. Pathol. 2017, 54, 571–578. [Google Scholar] [CrossRef]
- Riggio, M.; Perrone, M.C.; Polo, M.L.; Rodriguez, M.J.; May, M.; Abba, M.; Lanari, C.; Novaro, V. AKT1 and AKT2 Isoforms Play Distinct Roles during Breast Cancer Progression through the Regulation of Specific Downstream Proteins. Sci. Rep. 2017, 7, 44244. [Google Scholar] [CrossRef] [Green Version]
- Mayer, I.A.; Abramson, V.G.; Formisano, L.; Balko, J.M.; Estrada, M.V.; Sanders, M.E.; Juric, D.; Solit, D.; Berger, M.F.; Won, H.H.; et al. A Phase Ib Study of Alpelisib (BYL719), a PI3Kα-Specific Inhibitor, with Letrozole in ER+/HER2− Metastatic Breast Cancer. Clin. Cancer Res. 2017, 23, 26–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Port Louis, L.R.; Varshney, K.C.; Nair, M.G. An Immunohistochemical Study on the Expression of Sex Steroid Receptors in Canine Mammary Tumors. ISRN Vet. Sci. 2012, 2012, 378607. [Google Scholar] [CrossRef] [Green Version]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef]

| Grade | Histological Type | Number of Samples |
|---|---|---|
| Normal | N/A | 13 |
| Adenoma | Ductal adenoma | 12 |
| Intraductal papillary adenoma | 2 | |
| Complex adenoma | 10 | |
| Benign mixed tumor | 1 | |
| Low-grade carcinoma | Simple carcinoma-tubulopapillary | 1 |
| Simple carcinoma-tubular | 2 | |
| Complex carcinoma | 17 | |
| Carcinoma arising in a benign mixed tumor | 1 | |
| High-grade carcinoma | Simple carcinoma-solid | 17 |
| Simple carcinoma-tubulopapillary | 2 | |
| Simple carcinoma-tubular | 1 | |
| Simple carcinoma-comedocarcinoma | 2 | |
| Complex carcinoma | 1 | |
| Carcinosarcoma | 1 |
| Grade | Percentage (Number of Samples) | |
|---|---|---|
| PIK3CA-wild-type | 88% (73/83) | |
| PIK3CA-mutant | Normal (n = 13) | 0% (0/83) |
| Adenoma (n = 25) | 4.8% (4/83) | |
| Low-grade carcinoma (n = 21) | 6% (5/83) | |
| High-grade carcinoma (n = 24) | 1.2% (1/83) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.-H.; Seung, B.-J.; Cho, S.-H.; Lim, H.-Y.; Bae, M.-K.; Sur, J.-H. Dysregulation of PI3K/Akt/PTEN Pathway in Canine Mammary Tumor. Animals 2021, 11, 2079. https://doi.org/10.3390/ani11072079
Kim S-H, Seung B-J, Cho S-H, Lim H-Y, Bae M-K, Sur J-H. Dysregulation of PI3K/Akt/PTEN Pathway in Canine Mammary Tumor. Animals. 2021; 11(7):2079. https://doi.org/10.3390/ani11072079
Chicago/Turabian StyleKim, Soo-Hyeon, Byung-Joon Seung, Seung-Hee Cho, Ha-Young Lim, Min-Kyung Bae, and Jung-Hyang Sur. 2021. "Dysregulation of PI3K/Akt/PTEN Pathway in Canine Mammary Tumor" Animals 11, no. 7: 2079. https://doi.org/10.3390/ani11072079





