Rumen Bacterial Community of Grazing Lactating Yaks (Poephagus grunniens) Supplemented with Concentrate Feed and/or Rumen-Protected Lysine and Methionine
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Animals, Diet, Experimental Design, and Management
2.3. Sample Collection
2.4. Feed Analyses
2.5. Rumen Fermentation Variables
2.6. DNA Extraction, 16S rRNA Gene Amplification, and Sequencing
2.7. Statistical Analyses
3. Results
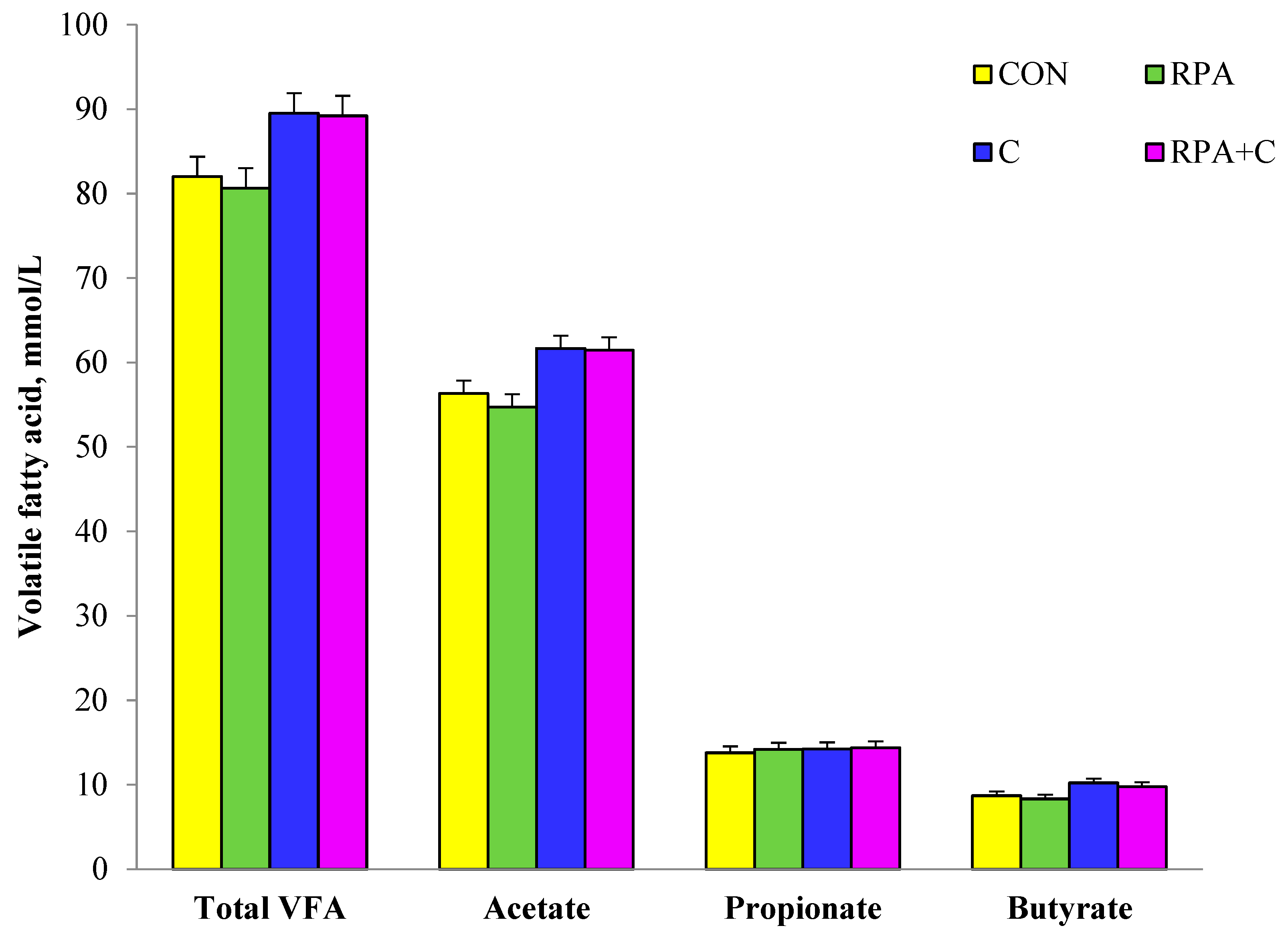
3.1. Rumen Fermentation Parameters
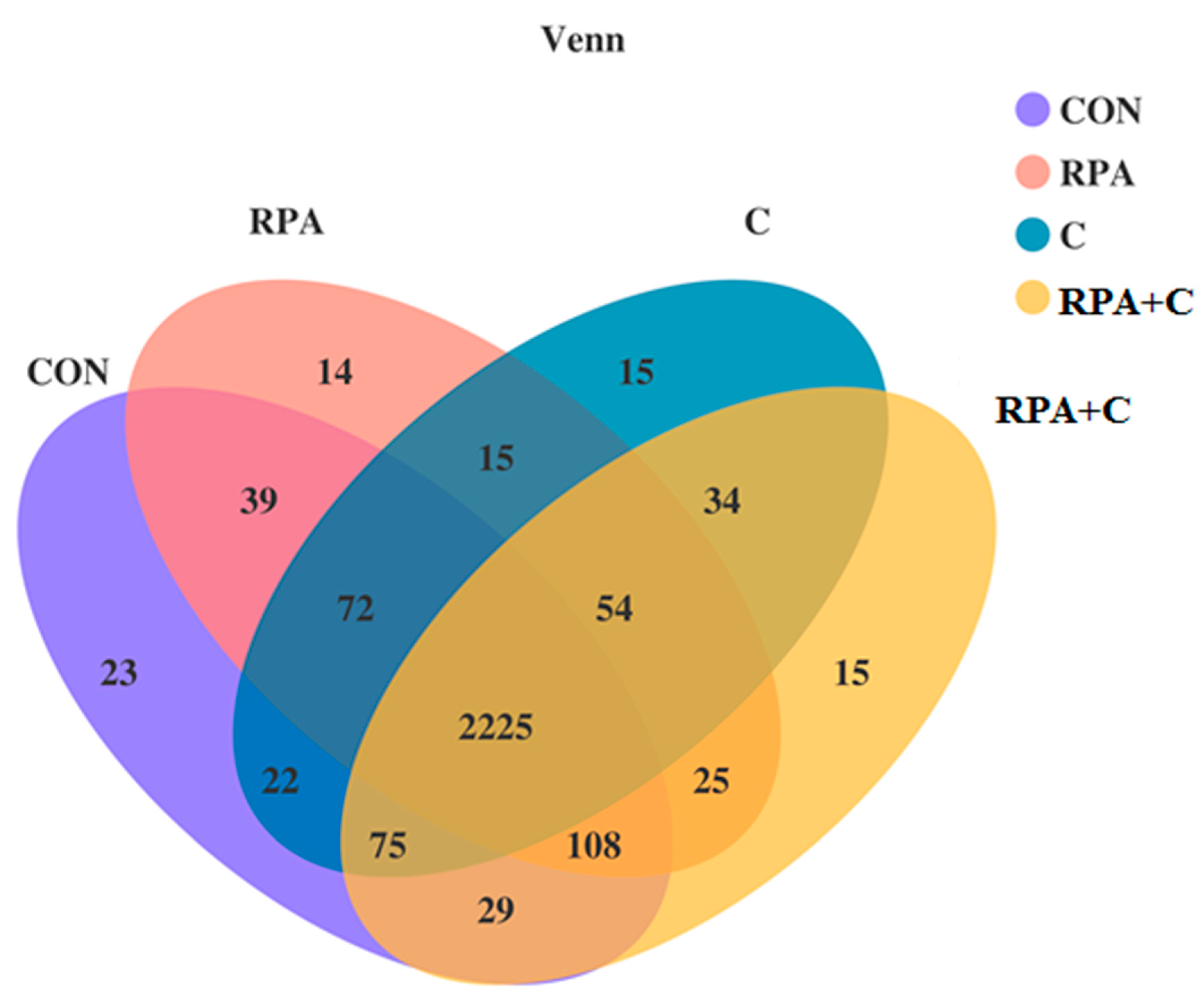
3.2. Collective Sequencing Data Summary
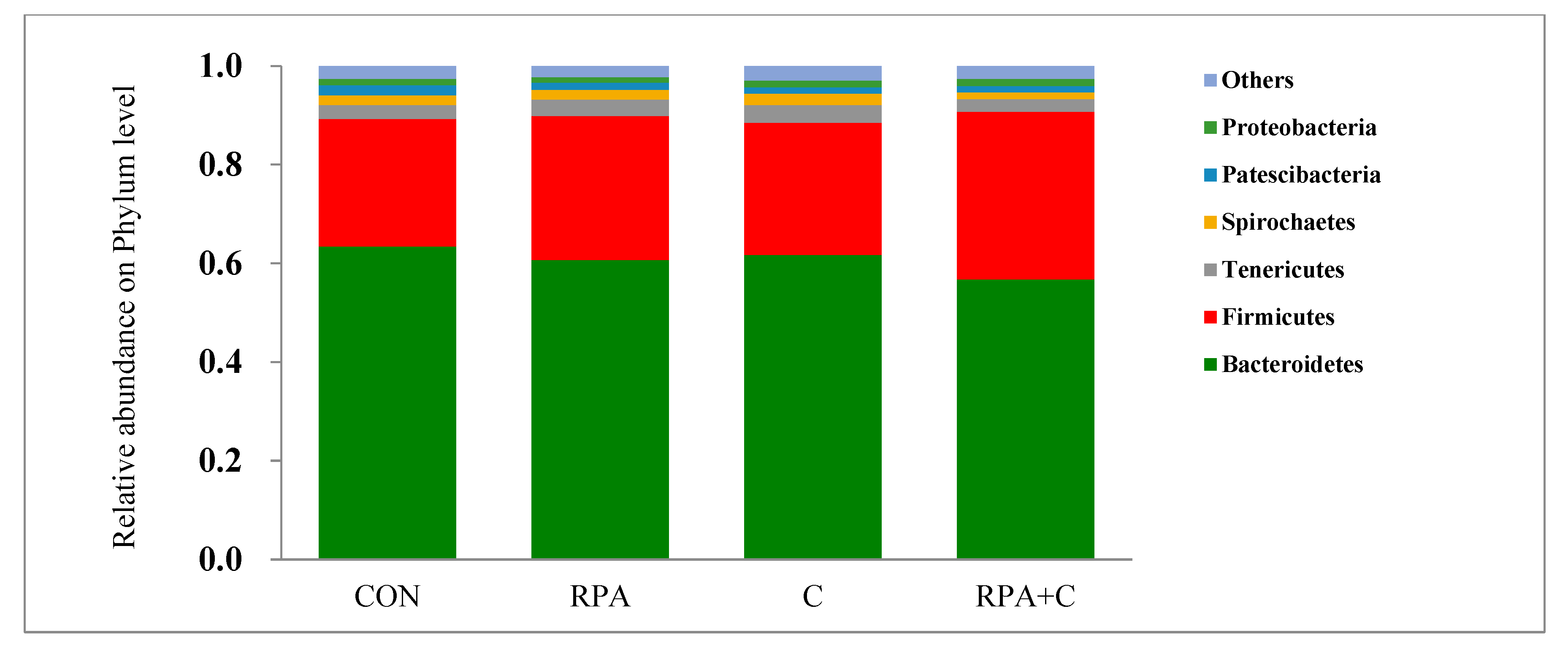
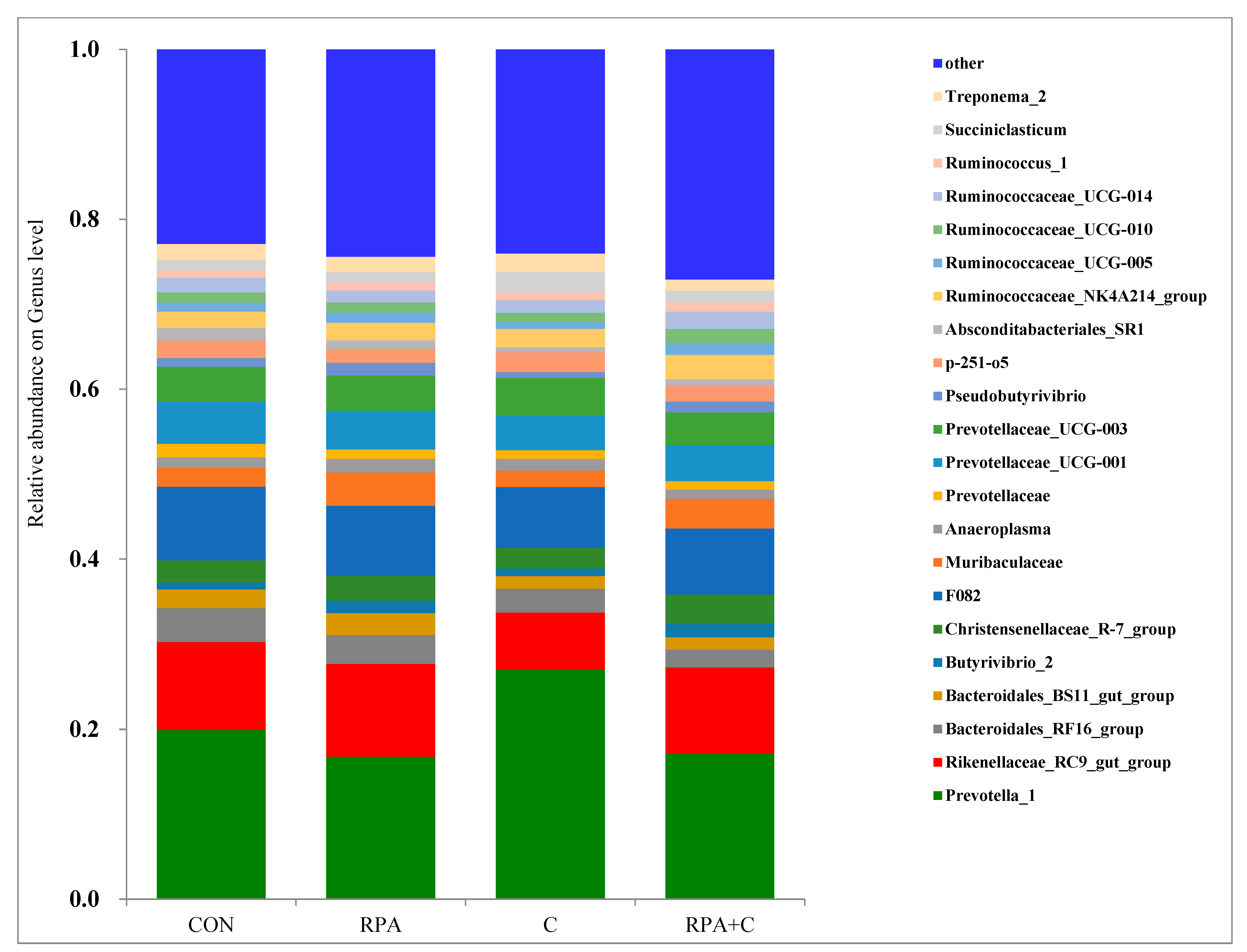
3.3. Bacterial Community Composition in the Rumen Fluid
4. Discussion
4.1. Rumen Fermentation Parameters
4.2. Effects of Supplementary Feeds on the Bacterial Community of the Rumen Fluid
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Guo, W.; Yang, L.; Wang, L.Z.; Wang, J.W.; Xu, Q.; Yan, T.H.; Xue, B. Evaluation of composition and individual variability of rumen microbiota in yaks by 16S rRNA high-throughput sequencing technology. Anaerobe 2015, 34, 4–9. [Google Scholar] [CrossRef]
- Hao, L.Z.; Xiang, Y.; Degen, A.; Huang, Y.Y.; Niu, J.Z.; Sun, L.; Chai, S.T.; Zhou, J.W.; Ding, L.M.; Long, R.J.; et al. Adding heat-treated rapeseed to the diet of yak improves growth performance and tenderness and nutritional quality of the meat. Anim. Sci. J. 2019, 90, 1177–1184. [Google Scholar] [CrossRef]
- Mi, J.D.; Zhou, J.W.; Ding, L.M.; Wang, L.; Long, R.J. Changes in the composition of yak colostrum during the first week of lactation. J. Dairy Sci. 2016, 99, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Fan, Q.S.; Wanapat, M.; Hou, F.J. Chemical Composition of milk and rumen microbiome diversity of yak, impacting by herbage grown at different phenological periods on the Qinghai-Tibet Plateau. Animals 2020, 10, 1030. [Google Scholar] [CrossRef]
- Bai, Y.F.; Rafiq, M.K.; Li, S.S.; Degen, A.A.; Mašek, O.; Sun, H.W.; Han, H.W.; Wang, T.; Joseph, S.; Bachmann, R.T.; et al. Biochar from pyrolyzed Tibetan Yak dung as a novel additive in ensiling sweet sorghum: An alternate to the hazardous use of yak dung as a fuel in the home. J. Hazard. Mater. 2021, 403, 123647. [Google Scholar] [CrossRef]
- Joshi, S.; Shrestha, L.; Bisht, N.; Wu, N.; Ismail, M.; Dorji, T.; Dangol, G.; Long, R.J. Ethnic and cultural diversity amongst yak herding communities in the Asian highlands. Sustainability 2020, 12, 957. [Google Scholar] [CrossRef]
- Ding, L.M.; Wang, Y.P.; Kreuzer, M.; Guo, X.S.; Mi, J.D.; Gou, Y.J.; Shang, Z.H.; Zhang, Y.; Zhou, J.W.; Wang, H.C.; et al. Seasonal variations in the fatty acid profile of milk from yaks grazing on the Qinghai-Tibetan plateau. J. Dairy Res. 2013, 80, 410–417. [Google Scholar] [CrossRef] [Green Version]
- Long, R.J. Seasonal Dynamics of Several Nutrient Metabolites in Serum of Alpine Grassland Grazing Yaks. Ph.D. Thesis, Gansu Agricultural University, Lanzhou, China, 1995; pp. 18–21. (In Chinese). [Google Scholar]
- Wang, H.C.; Zhao, Z.W.; Zhou, E.G. A preliminary study on amino acid nutrition requirements of lactating yak grazing in a warm season pasture in alpine meadows. Pratacult. Sci. 2019, 36, 1897–1907. (In Chinese) [Google Scholar]
- Russell, J.B.; Rychlik, J.L. Factors that alter rumen microbial ecology. Science 2001, 292, 1119–1122. [Google Scholar] [CrossRef]
- Martinez-Fernandez, G.; Jiao, J.; Padmanabha, J.; Denman, S.E.; McSweeney, C.S. Seasonal and nutrient supplement responses in rumen microbiota structure and metabolites of tropical rangeland cattle. Microorganisms 2020, 8, 1550. [Google Scholar] [CrossRef]
- Kong, F.L.; Gao, Y.X.; Tang, M.Q.; Fu, T.; Diao, Q.Y.; Bi, Y.L.; Tu, Y. Effects of dietary rumen-protected Lys levels on rumen fermentation and bacterial community composition in Holstein heifers. Appl. Microbiol. Biotechnol. 2020, 104, 6623–6634. [Google Scholar] [CrossRef]
- da Silva-Marques, R.P.; Zervoudakis, J.T.; Nakazato, L.; Hatamoto-Zervoudakis, L.K.; da Silva Cabral, L.; do Nascimento Matos, N.B.; da Silva, M.I.L.; Feliciano, A.L. Ruminal microbial populations and fermentation characteristics in beef cattle grazing tropical forage in dry season and supplemented with different protein levels. Curr. Microbiol. 2019, 76, 270–278. [Google Scholar] [CrossRef]
- Abdelmegeid, M.K.; Elolimy, A.A.; Zhou, Z.; Lopreiato, V.; McCann, J.C.; Loor, J.J. Rumen-protected methionine during the peripartal period in dairy cows and its effects on abundance of major species of ruminal bacteria. J. Anim. Sci. Biotechnol. 2018, 9, 17. [Google Scholar] [CrossRef] [Green Version]
- Fleming, A.J.; Estes, K.A.; Choi, H.; Barton, B.A.; Zimmerman, C.A.; Hanigan, M.D. Assessing bioavailability of ruminally protected methionine and lysine prototypes. J. Dairy Sci. 2019, 102, 4014–4024. [Google Scholar] [CrossRef]
- Liu, H.; Yang, G.; Degen, A.; Ji, K.X.; Jiao, D.; Liang, Y.P.; Xiao, L.; Long, R.J.; Zhou, J.W. Effect of feed level and supplementary rumen protected lysine and methionine on growth performance, rumen fermentation, blood metabolites and nitrogen balance in growing Tan lambs fed low protein diets. Anim. Feed Sci. Technol. 2021, 279, 115024. [Google Scholar] [CrossRef]
- Ren, J.Z.; Jin, J.H. Observation on the grazing habits of yak herds. China Anim. Husb. Vet. Med. 1956, 2, 58–62. (In Chinese) [Google Scholar]
- Shen, J.S.; Chai, Z.; Song, L.J.; Liu, J.X.; Wu, Y.M. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J. Dairy Sci. 2012, 95, 5978–5984. [Google Scholar] [CrossRef] [Green Version]
- AOAC. Official Methods of Analysis, 16th ed.; Association of Official Agricultural Chemists: Arlington, VA, USA, 1990. [Google Scholar]
- Van Soest, P.J.; Robertson, J.B.; Lewis, B.A. Methods for dietary fiber, neutral detergent fiber, and non-starch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Robertson, J.B.; Van Soest, P.J. The detergent system of analysis and its application to human foods. In The Analysis of Dietary Fibre in Food; James, W.P., Theander, O., Eds.; Marcel Dekker: New York, NY, USA, 1981; pp. 23–158. [Google Scholar]
- Liu, C.; Wu, H.; Liu, S.J.; Chai, S.T.; Meng, Q.X.; Zhou, Z.M. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front. Microbiol. 2019, 10, 1116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giger-Reverdin, S.; Rigalma, K.; Desnoyers, M.; Sauvant, D.; Duvaux-Ponter, C. Effect of concentrate level on feeding behavior and rumen and blood parameters in dairy goats: Relationships between behavioral and physiological parameters and effect of between-animal variability. J. Dairy Sci. 2014, 97, 4367–4678. [Google Scholar] [CrossRef] [Green Version]
- Balcells, J.; Aris, A.; Serrano, A.; Seradj, A.R.; Crespo, J.; Devant, M. Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets. J. Anim. Sci. 2012, 90, 4975–4984. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; You, W.; Tan, X.W.; Liu, G.F.; Zhang, X.L.; Liu, X.M.; Wan, F.C.; Wei, C. Caffeic acid modulates methane production and rumen fermentation in an opposite way with high-forage or high-concentrate substrate in vitro. J. Sci. Food Agric. 2021, 101, 3013–3020. [Google Scholar] [CrossRef]
- Xin, J.W.; Chai, Z.X.; Zhang, C.F.; Zhang, Q.; Zhu, Y.; Cao, H.W.; Zhong, J.C.; Ji, Q.M. Comparing the microbial community in four stomach of dairy cattle, yellow cattle and three yak herds in Qinghai-Tibetan plateau. Front. Microbiol. 2019, 10, 1547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, F.Y.; Guo, N.; Degen, A.A.; Niu, J.H.; Wei, H.Y.; Jing, X.P.; Ding, L.M.; Shang, Z.H.; Long, R.J. Effects of level of feed intake and season on digestibility of dietary components, efficiency of microbial protein synthesis, rumen fermentation and ruminal microbiota in yaks. Anim. Feed Sci. Technol. 2020, 259, 114359. [Google Scholar] [CrossRef]
- Zhao, Z.W.; Ma, Z.Y.; Wang, H.C.; Zhang, C.F. Effects of rumen-protected methionine and lysine supplementation on milk yields and components, rumen fermentation, and the rumen microbiome in lactating yaks (Bos grunniens). Anim. Feed Sci. Technol. 2021, 277, 114972. [Google Scholar] [CrossRef]
- Anderson, C.L.; Schneider, C.J.; Erickson, G.E.; MacDonald, J.C.; Fernando, S.C. Rumen bacterial communities can be acclimated faster to high concentrate diets than currently implemented feedlot programs. J. Appl. Microbiol. 2016, 120, 588–599. [Google Scholar] [CrossRef]
- Gruninger, R.J.; Ribeiro, G.O.; Cameron, A.; McAllister, T.A. Invited review: Application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal 2019, 13, 1843–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, R.; Zou, H.W.; Wang, H.Z.; Wang, Z.S.; Wang, X.Y.; Ma, J.; Shah, A.M.; Peng, Q.H.; Xue, B.; Wang, L.Z.; et al. Dietary energy levels affect rumen bacterial populations that influence the intramuscular fat fatty acids of fattening yaks (Bos grunniens). Animals 2020, 10, 1474. [Google Scholar] [CrossRef]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Liu, J.X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Li, H.; Li, R.; Chen, H.J.; Gao, J.; Wang, Y.; Zhang, Y.F.; Qi, Z.L. Effect of different seasons (spring vs summer) on the microbiota diversity in the feces of dairy cows. Int. J. Biometeorol. 2020, 64, 345–354. [Google Scholar] [CrossRef]
- Thomas, M.; Webb, M.; Ghimire, S.; Blair, A.; Olson, K.; Fenske, G.J.; Fonder, A.T.; Christopher-Hennings, J.; Brake, D.; Scaria, J. Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci. Rep. 2017, 7, 12257. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.D.; Tan, H.Y.; Long, R.J.; Liang, J.B.; Wright, A.D. Comparison of methanogen diversity of yak (Bos grunniens) and cattle (Bos taurus) from the Qinghai-Tibetan plateau, China. BMC Microbiol. 2012, 12, 237. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.H.; Bian, G.R.; Zhu, W.Y.; Mao, S.Y. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015, 6, 167. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Zhang, L.L.; Jin, W.; Meng, Z.X.; Cheng, Y.F.; Wang, J.; Zhu, W.Y. Methane emission, rumen fermentation, and microbial community response to a nitrooxy compound in low-quality forage fed Hu sheep. Curr. Microbiol. 2019, 76, 435–441. [Google Scholar] [CrossRef]
- Mao, S.Y.; Zhang, M.L.; Liu, J.H.; Zhu, W.Y. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef] [Green Version]
- Kopecný, J.; Zorec, M.; Mrázek, J.; Kobayashi, Y.; Marinsek-Logar, R. Butyrivibrio hungatei sp. nov. and Pseudobutyrivibrio xylanivorans sp. nov., butyrate-producing bacteria from the rumen. Int. J. Syst. Evol. Mic. 2003, 53 Pt 1, 201–209. [Google Scholar] [CrossRef] [Green Version]
- Byrd, K.M.; Gulati, A.S. The “gum-gut” axis in inflammatory bowel diseases: A hypothesis-driven review of associations and advances. Front. Immunol. 2021, 12, 620124. [Google Scholar] [CrossRef] [PubMed]
- Apajalahti, J.; Vienola, K.; Raatikainen, K.; Holder, V.; Moran, C.A. Conversion of branched-chain amino acids to corresponding isoacids-An in vitro tool for estimating ruminal protein degradability. Front. Vet. Med. 2019, 6, 311. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Porsch, M.; Grosse, I.; Hoffmann, K.; Schaller, H.G.; Reichert, S. Comparison of the oral microbiome of patients with generalized aggressive periodontitis and periodontitis-free subjects. Arch. Oral. Biol. 2019, 99, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.F.; Xue, M.Y.; Liu, J.X. Composition of rumen bacterial community in dairy cows with different levels of somatic cell counts. Front. Microbiol. 2018, 9, 3217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.F.; Xue, M.Y.; Sun, H.Z.; Valencak, T.G.; Guan, L.L.; Liu, J.X. Rumen and hindgut bacteria are potential indicators for mastitis of mid-lactating Holstein dairy cows. Microorganisms 2020, 8, 2042. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Chen, H.; Chen, F.; He, Y.X.; Zhao, C.; Zhu, D.; Zeng, L.L.; Li, W. Analysis of the rumen bacteria and methanogenic archaea of yak (Bos grunniens) steers grazing on the Qinghai-Tibetan Plateau. Livest. Sci. 2016, 188, 61–71. [Google Scholar]
- Solden, L.M.; Hoyt, D.W.; Collins, W.B.; Plank, J.E.; Daly, R.A.; Hildebrand, E.; Beavers, T.J.; Wolfe, R.; Nicora, C.D.; Purvine, S.O.; et al. New roles in hemicellulosic sugar fermentation for the uncultivated Bacteroidetes family BS11. ISME J. 2017, 11, 691–703. [Google Scholar] [CrossRef] [PubMed]
| Items | Forage | Concentrate |
|---|---|---|
| DM, % | 96.0 | 96.0 |
| CP, % | 16.7 | 15.3 |
| NDF, % | 50.4 | 10.63 |
| ADF, % | 29.9 | 5.52 |
| EE, % | 2.46 | 2.32 |
| Ash, % | 10.04 | 2.91 |
| ME, MJ/kg 1 | 9.80 | 12.60 |
| Alpha Diversity Indices | CON | RPA | C | RPA+C | Overall Data 1 | SEM 2 | p-Values 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| RPA | C | RPA × C | |||||||
| Sobs | 1839 | 1827 | 1713 | 1792 | 1793 | 46.4 | 0.552 | 0.052 | 0.243 |
| Shannon | 6.23 | 6.22 | 6.02 | 6.26 | 6.18 | 0.044 | 0.195 | 0.352 | 0.163 |
| Simpson | 0.0047 | 0.0047 | 0.0054 | 0.0045 | 0.0048 | 0.0009 | 0.364 | 0.626 | 0.309 |
| ACE | 2117 | 2106 | 2003 | 2048 | 2069 | 21.3 | 0.705 | 0.046 | 0.472 |
| Chao | 2141 | 2129 | 2029 | 2077 | 2094 | 22.5 | 0.694 | 0.079 | 0.486 |
| Coverage, % | 99.1 | 99.1 | 99.0 | 99.1 | 99.1 | 0.0003 | 0.984 | 0.604 | 0.270 |
| Items | CON | RPA | C | RPA+C | Overall Data 1 | SEM 2 | p-Value 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| RPA | C | RPA × C | |||||||
| Bacteroidetes | 64.3 | 61.4 | 62.6 | 57.6 | 61.5 | 2.84 | 0.275 | 0.251 | 0.663 |
| Firmicutes | 26.2 | 29.5 | 27.2 | 34.4 | 29.3 | 2.80 | 0.164 | 0.150 | 0.322 |
| Tenericutes | 2.82 | 3.33 | 3.60 | 2.59 | 3.09 | 0.476 | 0.612 | 0.973 | 0.144 |
| Spirochaetes | 2.05 | 1.98 | 2.36 | 1.43 | 1.96 | 0.411 | 0.254 | 0.775 | 0.318 |
| Patescibacteria | 2.03 | 1.53 | 1.31 | 1.34 | 1.55 | 0.231 | 0.333 | 0.078 | 0.281 |
| Proteobacteria | 1.36 | 1.07 | 1.38 | 1.41 | 1.31 | 0.177 | 0.573 | 0.176 | 0.228 |
| Others | 2.59 | 2.30 | 2.98 | 2.65 | 2.63 | 0.221 | 0.273 | 0.043 | 0.925 |
| F:B 3 ratio | 0.41 | 0.48 | 0.43 | 0.60 | 0.48 | 0.010 | <0.001 | <0.001 | 0.026 |
| Items | CON | RPA | C | RPA+C | Overall Data 1 | SEM 2 | p-Values 3 | ||
|---|---|---|---|---|---|---|---|---|---|
| RPA | C | RPA × C | |||||||
| Absconditabacteriales_SR1 | 1.48 | 1.04 | 0.58 | 0.75 | 1.0 | 0.165 | 0.448 | 0.005 | 0.099 |
| Anaeroplasma | 1.25 | 1.62 | 1.42 | 1.07 | 1.3 | 0.341 | 0.964 | 0.594 | 0.315 |
| Bacteroidales_RF16_group | 3.96 | 3.42 | 2.87 | 2.09 | 3.1 | 0.399 | 0.169 | 0.006 | 0.728 |
| Bacteroidales_BS11_gut_group | 2.20 | 2.59 | 1.49 | 1.45 | 1.9 | 0.430 | 0.730 | 0.031 | 0.576 |
| Butyrivibrio_2 | 0.83 | 1.46 | 0.86 | 1.68 | 1.2 | 0.310 | 0.041 | 0.685 | 0.774 |
| Christensenellaceae_R-7_group | 2.58 | 2.91 | 2.52 | 3.39 | 2.9 | 0.363 | 0.152 | 0.543 | 0.437 |
| F082 | 8.64 | 8.22 | 7.20 | 7.71 | 7.6 | 0.082 | 0.856 | 0.279 | 0.520 |
| Muribaculaceae | 2.23 | 3.9 | 1.96 | 3.49 | 2.9 | 0.930 | 0.115 | 0.720 | 0.943 |
| Prevotella_1 | 20.0 | 16.8 | 27.5 | 17.2 | 20.4 | 3.28 | 0.095 | 0.196 | 0.244 |
| Prevotellaceae | 1.58 | 1.11 | 1.02 | 1.00 | 1.20 | 0.157 | 0.231 | 0.013 | 0.070 |
| Prevotellaceae_UCG-001 | 4.95 | 4.52 | 4.16 | 4.24 | 4.5 | 0.685 | 0.835 | 0.312 | 0.616 |
| Prevotellaceae_UCG-003 | 4.10 | 4.16 | 4.50 | 3.85 | 4.2 | 0.575 | 0.672 | 0.913 | 0.452 |
| Pseudobutyrivibrio | 1.05 | 1.53 | 0.69 | 1.29 | 1.1 | 0.199 | 0.022 | 0.158 | 0.763 |
| p-251-o5 | 2.05 | 1.59 | 2.37 | 1.85 | 2.0 | 0.304 | 0.220 | 0.202 | 0.902 |
| Rikenellaceae_RC9_gut_group | 10.3 | 10.9 | 6.79 | 10.1 | 9.5 | 0.86 | 0.065 | 0.017 | 0.108 |
| Ruminococcaceae_NK4A214_group | 1.91 | 2.06 | 2.23 | 2.89 | 2.3 | 0.399 | 0.419 | 0.086 | 0.419 |
| Ruminococcaceae_UCG-005 | 1.08 | 1.20 | 0.80 | 1.39 | 1.1 | 0.172 | 0.085 | 0.756 | 0.160 |
| Ruminococcaceae_UCG-010 | 1.20 | 1.22 | 1.09 | 1.68 | 1.3 | 0.191 | 0.171 | 0.317 | 0.124 |
| Ruminococcaceae_UCG-014 | 1.73 | 1.37 | 1.53 | 2.01 | 1.7 | 0.336 | 0.886 | 0.380 | 0.119 |
| Ruminococcus_1 | 0.81 | 0.94 | 0.92 | 1.10 | 0.9 | 0.108 | 0.192 | 0.229 | 0.823 |
| Succiniclasticum | 1.26 | 1.23 | 2.41 | 1.37 | 1.6 | 0.454 | 0.269 | 0.181 | 0.294 |
| Treponema_2 | 1.90 | 1.82 | 2.24 | 1.31 | 1.8 | 0.403 | 0.240 | 0.834 | 0.314 |
| other | 22.9 | 24.4 | 24.4 | 27.1 | 24.7 | 1.27 | 0.198 | 0.063 | 0.563 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Jiang, H.; Hao, L.; Cao, X.; Degen, A.; Zhou, J.; Zhang, C. Rumen Bacterial Community of Grazing Lactating Yaks (Poephagus grunniens) Supplemented with Concentrate Feed and/or Rumen-Protected Lysine and Methionine. Animals 2021, 11, 2425. https://doi.org/10.3390/ani11082425
Liu H, Jiang H, Hao L, Cao X, Degen A, Zhou J, Zhang C. Rumen Bacterial Community of Grazing Lactating Yaks (Poephagus grunniens) Supplemented with Concentrate Feed and/or Rumen-Protected Lysine and Methionine. Animals. 2021; 11(8):2425. https://doi.org/10.3390/ani11082425
Chicago/Turabian StyleLiu, Hu, Hui Jiang, Lizhuang Hao, Xuliang Cao, Allan Degen, Jianwei Zhou, and Chengfu Zhang. 2021. "Rumen Bacterial Community of Grazing Lactating Yaks (Poephagus grunniens) Supplemented with Concentrate Feed and/or Rumen-Protected Lysine and Methionine" Animals 11, no. 8: 2425. https://doi.org/10.3390/ani11082425
APA StyleLiu, H., Jiang, H., Hao, L., Cao, X., Degen, A., Zhou, J., & Zhang, C. (2021). Rumen Bacterial Community of Grazing Lactating Yaks (Poephagus grunniens) Supplemented with Concentrate Feed and/or Rumen-Protected Lysine and Methionine. Animals, 11(8), 2425. https://doi.org/10.3390/ani11082425








