Using High-Resolution Differential Cell Counts (HRDCCs) in Bovine Milk and Blood to Monitor the Immune Status over the Entire Lactation Period
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Animal Study
2.2. Sampling, Cell Isolation, and Staining
2.3. Flow Cytometric Analysis and Gating
2.4. External Analysis of Blood Samples
3. Statistical Analysis
4. Results
4.1. Comparison to Automated Gating
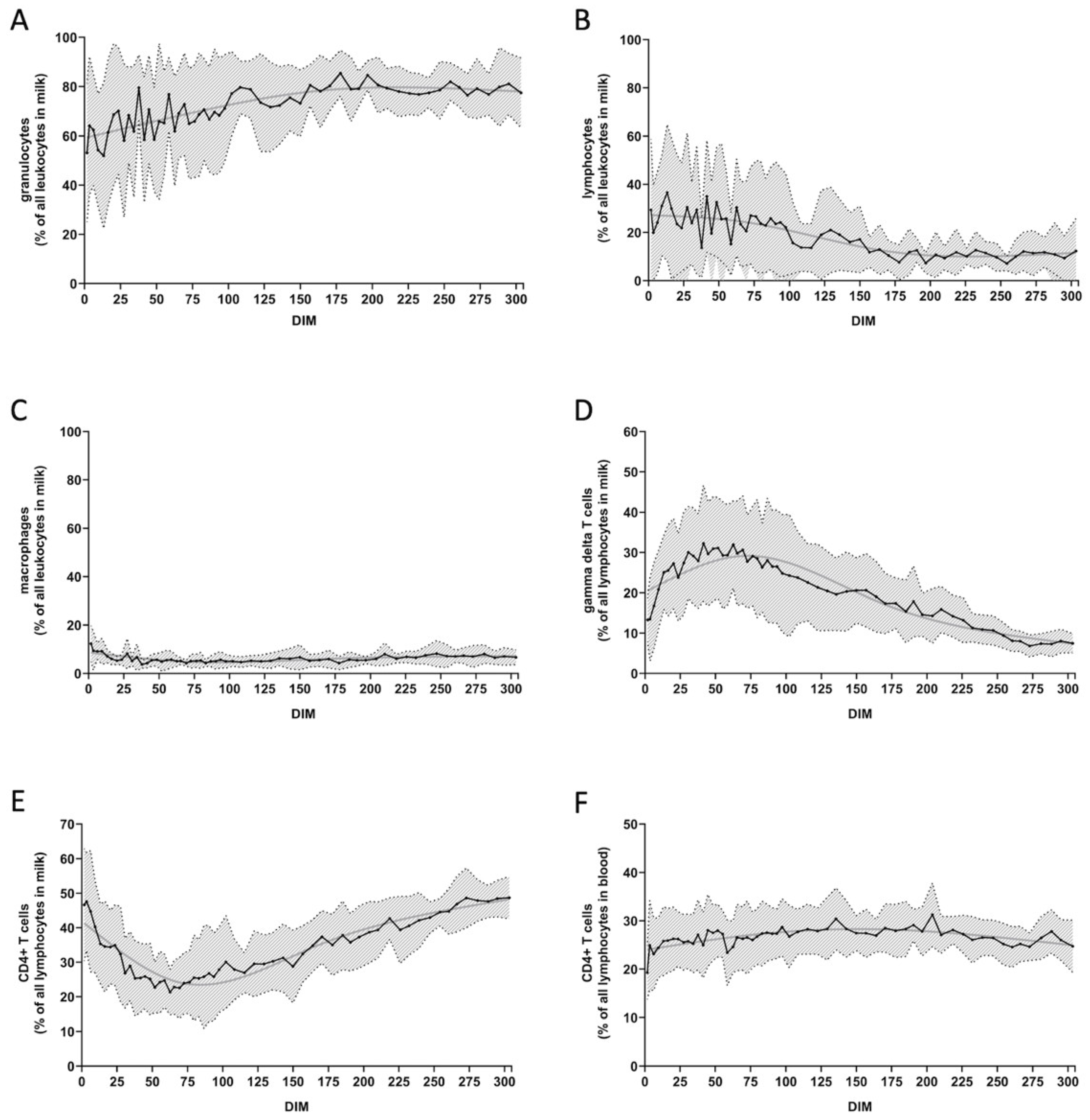
4.2. Progression of Cell Counts over Time
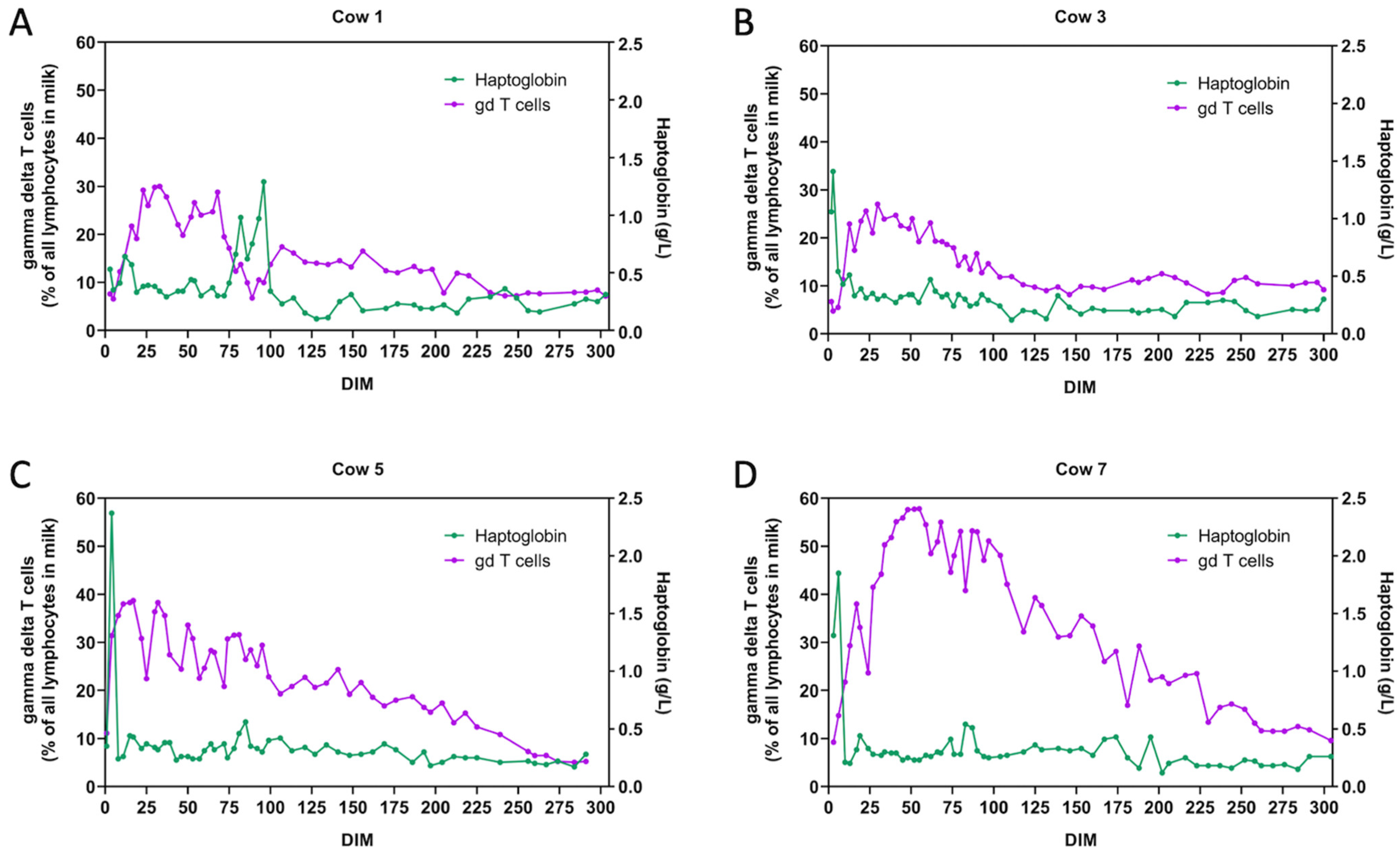
4.3. Deviations in Cell Counts in Case of Disease
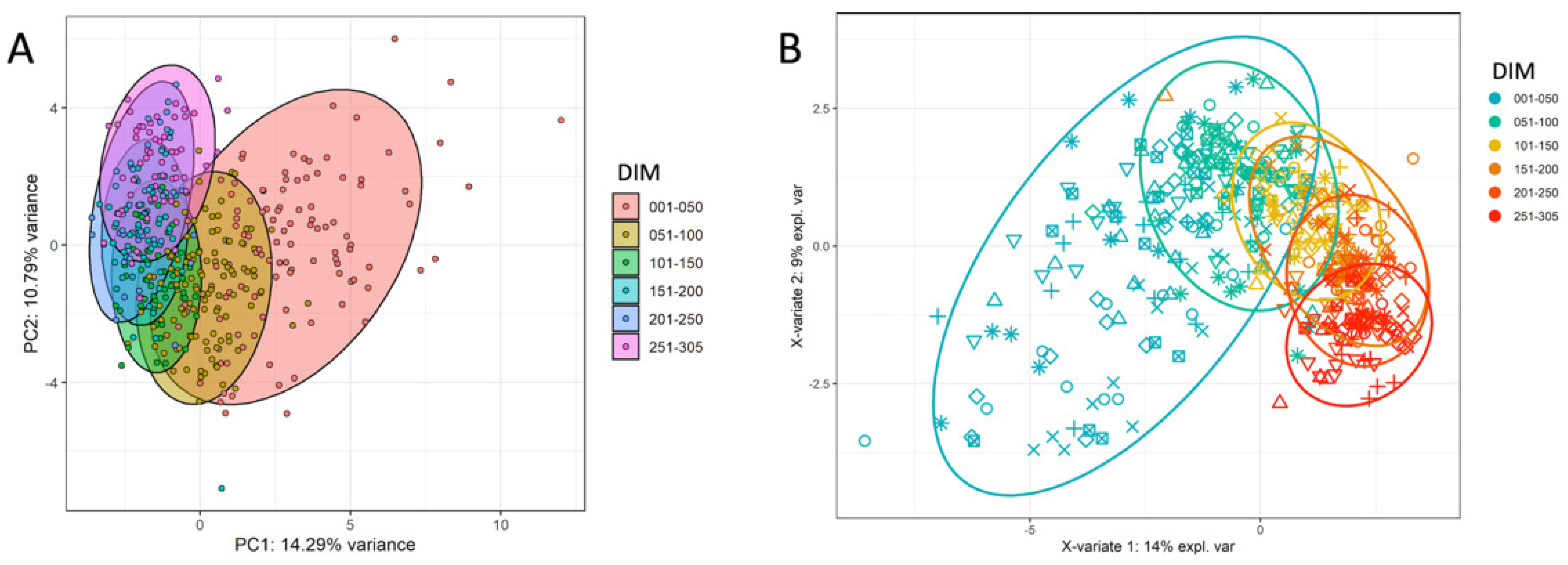
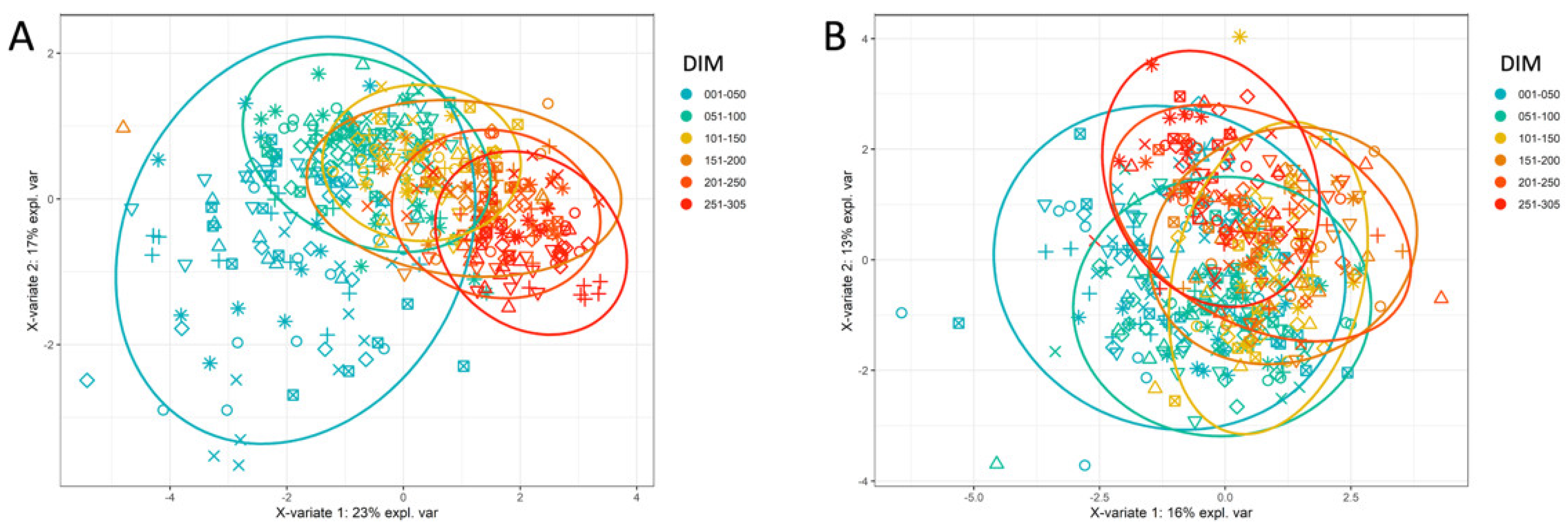
4.4. Unsupervised and Supervised Clustering
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barkema, H.W.; von Keyserlingk, M.A.; Kastelic, J.P.; Lam, T.J.; Luby, C.; Roy, J.P.; LeBlanc, S.J.; Keefe, G.P.; Kelton, D.F. Invited review: Changes in the dairy industry affecting dairy cattle health and welfare. J. Dairy Sci. 2015, 98, 7426–7445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buller, H.; Blokhuis, H.; Jensen, P.; Keeling, L. Towards Farm Animal Welfare and Sustainability. Animals 2018, 8, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weary, D.M.; von Keyserlingk, M.A.G. Public concerns about dairy-cow welfare: How should the industry respond? Anim. Prod. Sci. 2017, 57, 1201–1209. [Google Scholar] [CrossRef] [Green Version]
- Halasa, T.; Huijps, K.; Osteras, O.; Hogeveen, H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007, 29, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, F.J.; Santman-Berends, I.M.; Lam, T.J.; Hogeveen, H. Failure and preventive costs of mastitis on Dutch dairy farms. J. Dairy Sci. 2016, 99, 8365–8374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Borne, B.H.; Halasa, T.; van Schaik, G.; Hogeveen, H.; Nielen, M. Bioeconomic modeling of lactational antimicrobial treatment of new bovine subclinical intramammary infections caused by contagious pathogens. J. Dairy Sci. 2010, 93, 4034–4044. [Google Scholar] [CrossRef] [Green Version]
- Kuipers, A.; Koops, W.J.; Wemmenhove, H. Antibiotic use in dairy herds in the Netherlands from 2005 to 2012. J. Dairy Sci. 2016, 99, 1632–1648. [Google Scholar] [CrossRef] [Green Version]
- Prescott, J.F. The resistance tsunami, antimicrobial stewardship, and the golden age of microbiology. Vet. Microbiol. 2014, 171, 273–278. [Google Scholar] [CrossRef]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [Green Version]
- Adkins, P.R.F.; Middleton, J.R. Methods for Diagnosing Mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 479–491. [Google Scholar] [CrossRef]
- Dalen, G.; Rachah, A.; Norstebo, H.; Schukken, Y.H.; Reksen, O. The detection of intramammary infections using online somatic cell counts. J. Dairy Sci. 2019, 102, 5419–5429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, S.; Buckley, F.; Berry, D.P.; Rath, M.; Pierce, K.; Byrne, N.; Dillon, P. Effects of breed, feeding system, and parity on udder health and milking characteristics. J. Dairy Sci. 2007, 90, 5767–5779. [Google Scholar] [CrossRef] [PubMed]
- Schepers, A.J.; Lam, T.J.; Schukken, Y.H.; Wilmink, J.B.M.; Hanekamp, W.J.A. Estimation of Variance Components for Somatic Cell Counts to Determine Thresholds for Uninfected Quarters. J. Dairy Sci. 1997, 80, 1833–1840. [Google Scholar] [CrossRef]
- Oviedo-Boyso, J.; Valdez-Alarcon, J.J.; Cajero-Juarez, M.; Ochoa-Zarzosa, A.; Lopez-Meza, J.E.; Bravo-Patino, A.; Baizabal-Aguirre, V.M. Innate immune response of bovine mammary gland to pathogenic bacteria responsible for mastitis. J. Infect. 2007, 54, 399–409. [Google Scholar] [CrossRef]
- Sordillo, L.M. Mammary Gland Immunobiology and Resistance to Mastitis. Vet. Clin. N. Am. Food Anim. Pract. 2018, 34, 507–523. [Google Scholar] [CrossRef]
- Pilla, R.; Malvisi, M.; Snel, G.G.; Schwarz, D.; Konig, S.; Czerny, C.P.; Piccinini, R. Differential cell count as an alternative method to diagnose dairy cow mastitis. J. Dairy Sci. 2013, 96, 1653–1660. [Google Scholar] [CrossRef] [Green Version]
- Rivas, A.L.; Quimby, F.A.; Blue, J.; Coksaygan, O. Longitudinal evaluation of bovine mammary gland health status by somatic cell counting, flow cytometry, and cytology. J. Vet. Diagn. Investig. 2001, 13, 399–407. [Google Scholar] [CrossRef] [Green Version]
- Souza, F.N.; Blagitz, M.G.; Batista, C.F.; Takano, P.V.; Gargano, R.G.; Diniz, S.A.; Silva, M.X.; Ferronatto, J.A.; Santos, K.R.; Heinemann, M.B.; et al. Immune response in nonspecific mastitis: What can it tell us? J. Dairy Sci. 2020, 103, 5376–5386. [Google Scholar] [CrossRef]
- Farschtschi, S.; Mattes, M.; Hildebrandt, A.; Chiang, D.; Kirchner, B.; Kliem, H.; Pfaffl, M.W. Development of an advanced flow cytometry based high-resolution immunophenotyping method to benchmark early immune response in dairy cows. Sci. Rep. 2021, 11, 22896. [Google Scholar] [CrossRef]
- Schwarz, D.; Kleinhans, S.; Reimann, G.; Stückler, P.; Reith, F.; Ilves, K.; Pedastsaar, K.; Yan, L.; Zhang, Z.; Valdivieso, M.; et al. Investigation of dairy cow performance in different udder health groups defined based on a combination of somatic cell count and differential somatic cell count. Prev. Vet. Med. 2020, 183, 105123. [Google Scholar] [CrossRef]
- Finak, G.; Frelinger, J.; Jiang, W.; Newell, E.W.; Ramey, J.; Davis, M.M.; Kalams, S.A.; De Rosa, S.C.; Gottardo, R. OpenCyto: An open source infrastructure for scalable, robust, reproducible, and automated, end-to-end flow cytometry data analysis. PLoS Comput. Biol. 2014, 10, e1003806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finak, G.; Jiang, M. flowWorkspace: Infrastructure for Representing and Interacting with Gated and Ungated Cytometry Data Sets, R package version 3.33.7; 2019. [Google Scholar]
- Loken, M.R.; Brosnan, J.M.; Bach, B.A.; Ault, K.A. Establishing Optimal Lymphocyte Gates for Immunophenotyping by Flow Cytometry. Cytometry 1990, 11, 453–459. [Google Scholar] [CrossRef]
- Mayeno, A.N.; Hamann, K.J.; Gleich, G.J. Granule-associated flavin adenine dinucleotide (FAD) is responsible for eosinophil autofluorescence. J. Leukoc. Biol. 1992, 51, 172–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Merris, V.; Meyer, E.; Burvenich, C. Functional Maturation during Bovine Granulopoiesis. J. Dairy Sci. 2002, 85, 2859–2868. [Google Scholar] [CrossRef] [Green Version]
- Hussen, J.; Duvel, A.; Sandra, O.; Smith, D.; Sheldon, I.M.; Zieger, P.; Schuberth, H.J. Phenotypic and functional heterogeneity of bovine blood monocytes. PLoS ONE 2013, 8, e71502. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Kassambara, A.; Mundt, F. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses; R Package Version 1.0.7; 2020. [Google Scholar]
- Rohart, F.; Gautier, B.; Singh, A.; Le Cao, K.A. mixOmics: An R package for ′omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, D.; Rivas, A.L.; Konig, S.; Diesterbeck, U.S.; Schlez, K.; Zschock, M.; Wolter, W.; Czerny, C.P. CD2/CD21 index: A new marker to evaluate udder health in dairy cows. J. Dairy Sci. 2013, 96, 5106–5119. [Google Scholar] [CrossRef]
- Damm, M.; Holm, C.; Blaabjerg, M.; Bro, M.N.; Schwarz, D. Differential somatic cell count-A novel method for routine mastitis screening in the frame of Dairy Herd Improvement testing programs. J. Dairy Sci. 2017, 100, 4926–4940. [Google Scholar] [CrossRef] [Green Version]
- Halasa, T.; Kirkeby, C. Differential Somatic Cell Count: Value for Udder Health Management. Front. Vet. Sci. 2020, 7, 609055. [Google Scholar] [CrossRef]
- Yang, T.J.; Ayoub, I.A.; Rewinski, M.J. Lactation Stage-Dependent Changes of Lymphocyte Subpopulations in Mammary Secretions: Inversion of CD4+/CD8+ T Cell Ratios at Parturition. Am. J. Reprod. Immunol. 1997, 37, 378–383. [Google Scholar] [CrossRef]
- Asai, K.; Kai, K.; Rikiishi, H.; Sugawara, S.; Maruyama, Y.; Yamaguchi, T.; Ohta, M.; Kumagai, K. Variation in CD4+ T and CD8+ T lymphocyte subpopulations in bovine mammary gland secretions during lactating and non-lactating periods. Vet. Immunol. Immunopathol. 1998, 65, 51–61. [Google Scholar] [CrossRef]
- Hein, W.R.; Mackay, C.R. Prominence of gamma delta T cells in the ruminant immune system. Immunol. Today 1991, 12, 30–34. [Google Scholar] [CrossRef]
- Hoek, A.; Rutten, V.P.; Kool, J.; Arkesteijn, G.J.; Bouwstra, R.J.; Van Rhijn, I.; Koets, A.P. Subpopulations of bovine WC1(+) gammadelta T cells rather than CD4(+)CD25(high) Foxp3(+) T cells act as immune regulatory cells ex vivo. Vet. Res. 2009, 40, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman, E.; Hope, J.; Taylor, G.; Smith, A.L.; Cubillos-Zapata, C.; Charleston, B. Bovine gammadelta T cells are a major regulatory T cell subset. J. Immunol. 2014, 193, 208–222. [Google Scholar] [CrossRef] [Green Version]
- de Vries, M.J.; Veerkamp, R.F. Energy balance of dairy cattle in relation to milk production variables and fertility. J. Dairy Sci. 2000, 83, 62–69. [Google Scholar] [CrossRef]
- Ingvartsen, K.L.; Moyes, K. Nutrition, immune function and health of dairy cattle. Animal 2013, 7 (Suppl. S1), 112–122. [Google Scholar] [CrossRef] [Green Version]
- Harp, J.A.; Waters, T.E.; Goff, J.P. Lymphocyte subsets and adhesion molecule expression in milk and blood of periparturient dairy cattle. Vet. Immunol. Immunopathol. 2004, 102, 9–17. [Google Scholar] [CrossRef]
- Li, N.; Richoux, R.; Perruchot, M.H.; Boutinaud, M.; Mayol, J.F.; Gagnaire, V. Flow Cytometry Approach to Quantify the Viability of Milk Somatic Cell Counts after Various Physico-Chemical Treatments. PLoS ONE 2015, 10, e0146071. [Google Scholar] [CrossRef]
- Schröder, A.C.; Hamann, J. The influence of technical factors on differential cell count in milk. J. Dairy Res. 2005, 72, 153–158. [Google Scholar] [CrossRef]
- Sarikaya, H.; Werner-Misof, C.; Atzkern, M.; Bruckmaier, R.M. Distribution of leucocyte populations, and milk composition, in milk fractions of healthy quarters in dairy cows. J. Dairy Res. 2005, 72, 486–492. [Google Scholar] [CrossRef]
- Schwarz, D.; Kleinhans, S.; Reimann, G.; Stuckler, P.; Reith, F.; Ilves, K.; Pedastsaar, K.; Yan, L.; Zhang, Z.; Lorenzana, R.; et al. Associations between different udder health groups defined based on a combination of total and differential somatic cell count and the future udder health status of dairy cows. Prev. Vet. Med. 2021, 192, 105374. [Google Scholar] [CrossRef] [PubMed]
- Rivas, A.L.; Quimby, F.W.; Coksaygan, O.; Olmstead, L.; Lein, D.H. Longitudinal evaluation of CD4+ and CD8+ peripheral blood and mammary gland lymphocytes in cows experimentally inoculated with Staphylococcus aureus. Can. J. Vet. Res. 2000, 64, 232–237. [Google Scholar] [PubMed]
- Soltys, J.; Quinn, M.T. Selective recruitment of T-cell subsets to the udder during staphylococcal and streptococcal mastitis: Analysis of lymphocyte subsets and adhesion molecule expression. Infect. Immun. 1999, 67, 6293–6302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rainard, P.; Cunha, P.; Ledresseur, M.; Staub, C.; Touze, J.L.; Kempf, F.; Gilbert, F.B.; Foucras, G. Antigen-Specific Mammary Inflammation Depends on the Production of IL-17A and IFN-gamma by Bovine CD4+ T Lymphocytes. PLoS ONE 2015, 10, e0137755. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farschtschi, S.; Hildebrandt, A.; Mattes, M.; Kirchner, B.; Pfaffl, M.W. Using High-Resolution Differential Cell Counts (HRDCCs) in Bovine Milk and Blood to Monitor the Immune Status over the Entire Lactation Period. Animals 2022, 12, 1339. https://doi.org/10.3390/ani12111339
Farschtschi S, Hildebrandt A, Mattes M, Kirchner B, Pfaffl MW. Using High-Resolution Differential Cell Counts (HRDCCs) in Bovine Milk and Blood to Monitor the Immune Status over the Entire Lactation Period. Animals. 2022; 12(11):1339. https://doi.org/10.3390/ani12111339
Chicago/Turabian StyleFarschtschi, Sabine, Alex Hildebrandt, Martin Mattes, Benedikt Kirchner, and Michael W. Pfaffl. 2022. "Using High-Resolution Differential Cell Counts (HRDCCs) in Bovine Milk and Blood to Monitor the Immune Status over the Entire Lactation Period" Animals 12, no. 11: 1339. https://doi.org/10.3390/ani12111339






