Research Progress in Porcine Reproductive and Respiratory Syndrome Virus–Host Protein Interactions
Abstract
:Simple Summary
Abstract
1. Introduction
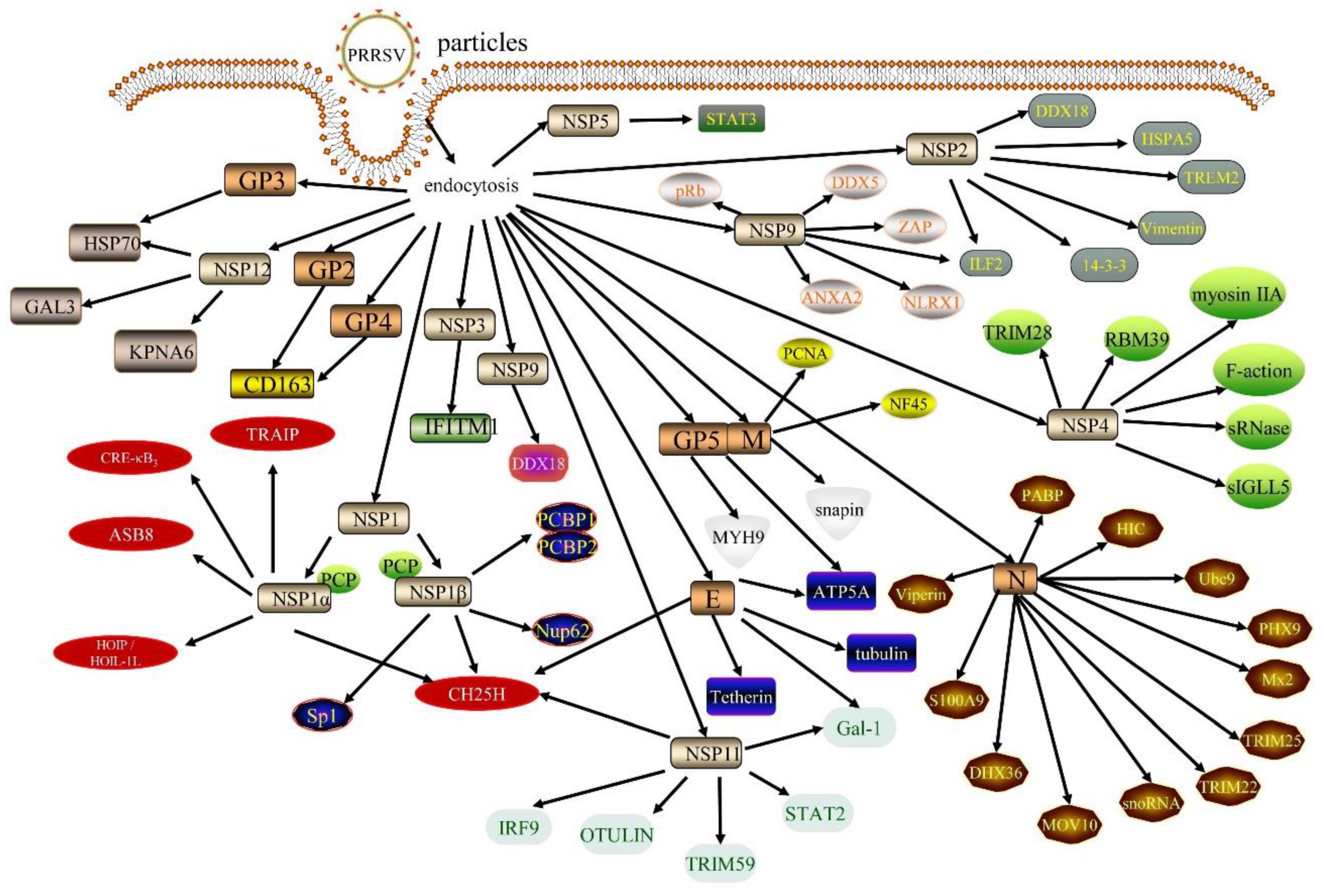
2. Nonstructural Proteins
2.1. NSP1
2.2. NSP2
2.3. NSP4
2.4. NSP5
2.5. NSP7
2.6. NSP9
2.7. NSP10
2.8. NSP11
2.9. NSP12
2.10. NSP3, 6, and 8
3. Structural Proteins
3.1. GP2
3.2. E
3.3. GP3
3.4. GP4
3.5. GP5a
3.6. GP5
3.7. M
3.8. N
4. Summaries
5. Perspectives
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rossow, K.D. Porcine reproductive and respiratory syndrome. Vet. Pathol. 1998, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Wensvoort, G.; Terpstra, C.; Pol, J.M.; ter Laak, E.A.; Bloemraad, M.; de Kluyver, E.P.; Kragten, C.; van Buiten, L.; den Besten, A.; Wagenaar, F.; et al. Mystery swine disease in The Netherlands: The isolation of Lelystad virus. Vet. Q. 1991, 13, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Benfield, D.A.; Nelson, E.; Collins, J.E.; Harris, L.; Goyal, S.M.; Robison, D.; Christianson, W.T.; Morrison, R.B.; Gorcyca, D.; Chladek, D. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332). J. Vet. Diagn. Investig. 1992, 4, 127–133. [Google Scholar] [CrossRef] [PubMed]
- An, T.Q.; Zhou, Y.J.; Liu, G.Q.; Tian, Z.J.; Li, J.; Qiu, H.J.; Tong, G.Z. Genetic diversity and phylogenetic analysis of glycoprotein 5 of PRRSV isolates in mainland China from 1996 to 2006: Coexistence of two NA-subgenotypes with great diversity. Vet. Microbiol. 2007, 123, 43–52. [Google Scholar] [CrossRef]
- Tian, K.; Yu, X.; Zhao, T.; Feng, Y.; Cao, Z.; Wang, C.; Hu, Y.; Chen, X.; Hu, D.; Tian, X.; et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ni, J.; Cao, Z.; Han, X.; Xia, Y.; Zi, Z.; Ning, K.; Liu, Q.; Cai, L.; Qiu, P.; et al. The epidemic status and genetic diversity of 14 highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV) isolates from China in 2009. Vet. Microbiol. 2011, 150, 257–269. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Z.; Ding, Y.; Ge, X.; Guo, X.; Yang, H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 2015, 21, 2256–2257. [Google Scholar] [CrossRef]
- Li, X.; Wu, J.; Tan, F.; Li, Y.; Ji, G.; Zhuang, J.; Zhai, X.; Tian, K. Genome characterization of two NADC30-like porcine reproductive and respiratory syndrome viruses in China. Springerplus 2016, 5, 1677. [Google Scholar] [CrossRef] [Green Version]
- Prieto, C.; Suarez, P.; Martin-Rillo, S.; Simarro, I.; Solana, A.; Castro, J.M. Effect of porcine reproductive and respiratory syndrome virus (PRRSV) on development of porcine fertilized ova in vitro. Theriogenology 1996, 46, 687–693. [Google Scholar] [CrossRef]
- Mardassi, H.; Mounir, S.; Dea, S. Molecular analysis of the ORFs 3 to 7 of porcine reproductive and respiratory syndrome virus, Quebec reference strain. Arch. Virol. 1995, 140, 1405–1418. [Google Scholar] [CrossRef]
- Kappes, M.A.; Faaberg, K.S. PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479–480, 475–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Xue, F.; Guo, Y.; Ma, M.; Hao, N.; Zhang, X.C.; Lou, Z.; Li, X.; Rao, Z. Crystal structure of porcine reproductive and respiratory syndrome virus leader protease Nsp1alpha. J. Virol. 2009, 83, 10931–10940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, W.; Fang, L.; Jing, H.; Tao, R.; Wang, T.; Li, Y.; Long, S.; Wang, D.; Xiao, S. Cholesterol 25-Hydroxylase Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication through Enzyme Activity-Dependent and -Independent Mechanisms. J. Virol. 2017, 91, e00827-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, P.; Su, Y.; Li, R.; Zhang, L.; Chen, C.; Zhang, L.; Faaberg, K.; Huang, J. Dual regulation of host TRAIP post-translation and nuclear/plasma distribution by porcine reproductive and respiratory syndrome virus non-structural protein 1alpha promotes viral proliferation. Front. Immunol. 2018, 9, 3023. [Google Scholar] [CrossRef]
- Jing, H.; Fang, L.; Ding, Z.; Wang, D.; Hao, W.; Gao, L.; Ke, W.; Chen, H.; Xiao, S. Porcine Reproductive and Respiratory Syndrome Virus nsp1alpha Inhibits NF-kappaB Activation by Targeting the Linear Ubiquitin Chain Assembly Complex. J. Virol. 2017, 91, e01911-16. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Chen, C.; He, J.; Zhang, L.; Zhang, L.; Guo, Y.; Zhang, W.; Tan, K.; Huang, J. E3 ligase ASB8 promotes porcine reproductive and respiratory syndrome virus proliferation by stabilizing the viral Nsp1alpha protein and degrading host IKKbeta kinase. Virology 2019, 532, 55–68. [Google Scholar] [CrossRef]
- Dong, H.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. Porcine reproductive and respiratory syndrome virus nsp1beta and nsp11 antagonize the antiviral activity of cholesterol-25-hydroxylase via lysosomal degradation. Vet. Microbiol. 2018, 223, 134–143. [Google Scholar] [CrossRef]
- Beura, L.K.; Dinh, P.X.; Osorio, F.A.; Pattnaik, A.K. Cellular poly(c) binding proteins 1 and 2 interact with porcine reproductive and respiratory syndrome virus nonstructural protein 1beta and support viral replication. J. Virol. 2011, 85, 12939–12949. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; He, Q.; Gao, Y.; Guo, X.; Ge, X.; Zhou, L.; Yang, H. Interaction of cellular poly(C)-binding protein 2 with nonstructural protein 1beta is beneficial to Chinese highly pathogenic porcine reproductive and respiratory syndrome virus replication. Virus Res. 2012, 169, 222–230. [Google Scholar] [CrossRef]
- Wang, R.; Nan, Y.; Yu, Y.; Zhang, Y.J. Porcine reproductive and respiratory syndrome virus Nsp1beta inhibits interferon-activated JAK/STAT signal transduction by inducing karyopherin-alpha1 degradation. J. Virol. 2013, 87, 5219–5228. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.; Nan, Y.; Shen, M.; Ritthipichai, K.; Zhu, X.; Zhang, Y.J. Porcine reproductive and respiratory syndrome virus inhibits type I interferon signaling by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2010, 84, 11045–11055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, H.; Han, M.; Kim, J.; Gustin, K.E.; Yoo, D. Porcine reproductive and respiratory syndrome virus nonstructural protein 1 beta interacts with nucleoporin 62 to promote viral replication and immune evasion. J. Virol. 2019, 93, e00469-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramaniam, S.; Kwon, B.; Beura, L.K.; Kuszynski, C.A.; Pattnaik, A.K.; Osorio, F.A. Porcine reproductive and respiratory syndrome virus non-structural protein 1 suppresses tumor necrosis factor-alpha promoter activation by inhibiting NF-kappaB and Sp1. Virology 2010, 406, 270–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Rutherford, M.S.; Faaberg, K.S. The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities. J. Virol. 2009, 83, 9449–9463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.; Zhou, L.; Ge, X.; Zhang, H.; Zhang, R.; Wang, C.; Wang, L.; Zhang, Z.; Yang, H.; Guo, X. Cellular DEAD-box RNA helicase 18 (DDX18) Promotes the PRRSV Replication via Interaction with Virus nsp2 and nsp10. Virus Res. 2017, 238, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Bian, T.; Zhang, Z.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H.; Yu, K. Interleukin-2 enhancer binding factor 2 interacts with the nsp9 or nsp2 of porcine reproductive and respiratory syndrome virus and exerts negatively regulatory effect on the viral replication. Virol. J. 2017, 14, 125. [Google Scholar] [CrossRef]
- Song, T.; Fang, L.; Wang, D.; Zhang, R.; Zeng, S.; An, K.; Chen, H.; Xiao, S. Quantitative interactome reveals that porcine reproductive and respiratory syndrome virus nonstructural protein 2 forms a complex with viral nucleocapsid protein and cellular vimentin. J. Proteom. 2016, 142, 70–81. [Google Scholar] [CrossRef]
- Xiao, Y.; Wu, W.; Gao, J.; Smith, N.; Burkard, C.; Xia, D.; Zhang, M.; Wang, C.; Archibald, A.; Digard, P.; et al. Characterization of the Interactome of the Porcine Reproductive and Respiratory Syndrome Virus Nonstructural Protein 2 Reveals the Hyper Variable Region as a Binding Platform for Association with 14-3-3 Proteins. J. Proteome Res. 2016, 15, 1388–1401. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Cong, F.; Tan, M.; Ding, G.; Liu, J.; Li, L.; Zhao, Y.; Liu, S.; Xiao, Y. 14-3-3epsilon acts as a proviral factor in highly pathogenic porcine reproductive and respiratory syndrome virus infection. Vet. Res. 2019, 50, 16. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Liu, J.; Ding, G.; Shao, Q.; Wang, B.; Li, Y.; Feng, J.; Zhao, Y.; Liu, S.; Xiao, Y. The tail domain of PRRSV NSP2 plays a key role in aggrephagy by interacting with 14-3-3epsilon. Vet. Res. 2020, 51, 104. [Google Scholar] [CrossRef]
- Han, J.; Rutherford, M.S.; Faaberg, K.S. Proteolytic products of the porcine reproductive and respiratory syndrome virus nsp2 replicase protein. J. Virol. 2010, 84, 10102–10112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Zhang, X.; Dong, W.; Wang, X.; He, S.; Zhang, H.; Wang, X.; Wei, R.; Chen, Y.; Liu, X.; et al. TREM2 suppresses the proinflammatory response to facilitate PRRSV infection via PI3K/NF-kappaB signaling. PLoS Pathog. 2020, 16, e1008543. [Google Scholar] [CrossRef]
- Sun, Z.; Li, Y.; Ransburgh, R.; Snijder, E.J.; Fang, Y. Nonstructural protein 2 of porcine reproductive and respiratory syndrome virus inhibits the antiviral function of interferon-stimulated gene 15. J. Virol. 2012, 86, 3839–3850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, X.; Lu, G.; Gao, F.; Peng, H.; Feng, Y.; Ma, G.; Bartlam, M.; Tian, K.; Yan, J.; Hilgenfeld, R.; et al. Structure and cleavage specificity of the chymotrypsin-like serine protease (3CLSP/nsp4) of porcine reproductive and respiratory syndrome virus (PRRSV). J. Mol. Biol. 2009, 392, 977–993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M. Prokaryotic Expression and Identification of Swine Ribonuclease L (sRNase L) and Its Interaction with PRRSV Proteins. Ph.D. Thesis, Shandong Agricultural University, Tai’an, China, 2018. [Google Scholar]
- Yin, M. sIGLL5, a Novel Adaptor of RIG-I and MDA5, Interacts with PRRSV Nsp4 and Inhibits Type I Interferon Production In Vitro. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2013. [Google Scholar]
- Chen, M. The Molecular Mechanisms of TRIM28 in Regulation of PRRSV-induced Autophagy. Ph.D. Thesis, Shandong Agricultural University, Tai’an, China, 2020. [Google Scholar] [CrossRef]
- Song, Y.; Guo, Y.; Li, X.; Sun, R.; Zhu, M.; Shi, J.; Tan, Z.; Zhang, L.; Huang, J. RBM39 Alters phosphorylation of c-jun and binds to viral RNA to promote PRRSV proliferation. Front. Immunol. 2021, 12, 664417. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Katz, B.B.; Tomich, J.M.; Gallagher, T.; Fang, Y. Porcine Reproductive and Respiratory Syndrome Virus Utilizes Nanotubes for Intercellular Spread. J. Virol 2016, 90, 5163–5175. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Wang, R.; Ma, Z.; Xiao, Y.; Nan, Y.; Wang, Y.; Lin, S.; Zhang, Y.J. Porcine reproductive and respiratory syndrome virus antagonizes JAK/STAT3 signaling via nsp5, which induces STAT3 degradation. J. Virol 2017, 91, e02087-16. [Google Scholar] [CrossRef] [Green Version]
- Van Aken, D.; Zevenhoven-Dobbe, J.; Gorbalenya, A.E.; Snijder, E.J. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 2006, 87, 3473–3482. [Google Scholar] [CrossRef]
- Chen, J.; Xu, X.; Tao, H.; Li, Y.; Nan, H.; Wang, Y.; Tian, M.; Chen, H. Structural analysis of porcine reproductive and respiratory syndrome virus non-structural protein 7alpha (NSP7alpha) and identification of its interaction with NSP9. Front. Microbiol. 2017, 8, 853. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Ge, X.; Wang, X.; Liu, A.; Guo, X.; Zhou, L.; Yu, K.; Yang, H. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res. 2015, 195, 217–224. [Google Scholar] [CrossRef]
- Dong, J.; Zhang, N.; Ge, X.; Zhou, L.; Guo, X.; Yang, H. The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology 2014, 464–465, 432–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Guo, D.; Huang, L.; Yin, M.; Liu, Q.; Wang, Y.; Yang, C.; Liu, Y.; Zhang, L.; Tian, Z.; et al. The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res. 2014, 189, 106–113. [Google Scholar] [CrossRef]
- Jing, H.; Song, T.; Cao, S.; Sun, Y.; Wang, J.; Dong, W.; Zhang, Y.; Ding, Z.; Wang, T.; Xing, Z.; et al. Nucleotide-binding oligomerization domain-like receptor X1 restricts porcine reproductive and respiratory syndrome virus-2 replication by interacting with viral Nsp9. Virus Res. 2019, 268, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Song, Z.; Bai, J.; Liu, X.; Nauwynck, H.; Jiang, P. ZAP, a CCCH-Type Zinc Finger Protein, Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication and Interacts with Viral Nsp9. J. Virol. 2019, 93, e00001-19. [Google Scholar] [CrossRef] [Green Version]
- Bautista, E.M.; Faaberg, K.S.; Mickelson, D.; McGruder, E.D. Functional properties of the predicted helicase of porcine reproductive and respiratory syndrome virus. Virology 2002, 298, 258–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wen, X.; Dong, J.; Ge, X.; Zhou, L.; Yang, H.; Guo, X. Epitope mapping and characterization of a novel Nsp10-specific monoclonal antibody that differentiates genotype 2 PRRSV from genotype 1 PRRSV. Virol. J. 2017, 14, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.; Yoo, D. The small envelope protein of porcine reproductive and respiratory syndrome virus possesses ion channel protein-like properties. Virology 2006, 355, 30–43. [Google Scholar] [CrossRef] [Green Version]
- Opriessnig, T.; Madson, D.M.; Prickett, J.R.; Kuhar, D.; Lunney, J.K.; Elsener, J.; Halbur, P.G. Effect of porcine circovirus type 2 (PCV2) vaccination on porcine reproductive and respiratory syndrome virus (PRRSV) and PCV2 coinfection. Vet. Microbiol. 2008, 131, 103–114. [Google Scholar] [CrossRef]
- Shi, X.; Zhang, X.; Chang, Y.; Jiang, B.; Deng, R.; Wang, A.; Zhang, G. Nonstructural protein 11 (nsp11) of porcine reproductive and respiratory syndrome virus (PRRSV) promotes PRRSV infection in MARC-145 cells. BMC Vet. Res. 2016, 12, 90. [Google Scholar] [CrossRef] [Green Version]
- Wen, X.; Ge, X.; Zhou, L.; Zhang, Y.; Guo, X.; Yang, H. PRRSV Promotes MARC-145 Cells Entry Into S Phase of the Cell Cycle to Facilitate Viral Replication via Degradation of p21 by nsp11. Front. Vet. Sci 2021, 8, 642095. [Google Scholar] [CrossRef]
- Wang, D.; Chen, J.; Yu, C.; Zhu, X.; Xu, S.; Fang, L.; Xiao, S. Porcine reproductive and respiratory syndrome virus nsp11 antagonizes type I interferon signaling by targeting IRF9. J. Virol. 2019, 93, e00623-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Y.; Shi, P.; Zhang, L.; Lu, D.; Zhao, C.; Li, R.; Zhang, L.; Huang, J. The superimposed deubiquitination effect of OTULIN and porcine reproductive and respiratory syndrome virus (PRRSV) Nsp11 promotes multiplication of PRRSV. J. Virol. 2018, 92, e00175-18. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Ke, W.; Tao, R.; Li, Y.; Zhao, Y.; Cao, S.; Sun, Y.; Wang, J.; Zhang, Y.; Dong, W.; et al. TRIM59 inhibits porcine reproductive and respiratory syndrome virus (PRRSV)-2 replication in vitro. Res. Vet. Sci. 2019, 127, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Ke, H.; Han, M.; Chen, N.; Fang, W.; Yoo, D. Nonstructural protein 11 of porcine reproductive and respiratory syndrome virus suppresses both MAVS and RIG-I expression as one of the mechanisms to antagonize type I interferon production. PLoS ONE 2016, 11, e0168314. [Google Scholar] [CrossRef]
- Li, L.; Zhao, K.; Gao, F.; Jiang, Y.; Shan, T.; Tong, W.; Zheng, H.; Yu, L.; Li, G.; Ma, Z.; et al. Restriction of porcine reproductive and respiratory syndrome virus replication by galectin-1. Vet. Microbiol. 2019, 235, 310–318. [Google Scholar] [CrossRef]
- Yang, L.; He, J.; Wang, R.; Zhang, X.; Lin, S.; Ma, Z.; Zhang, Y. Nonstructural protein 11 of porcine reproductive and respiratory syndrome virus induces STAT2 degradation to inhibit interferon signaling. J. Virol. 2019, 93, e01352-19. [Google Scholar] [CrossRef]
- Wang, T.Y.; Fang, Q.Q.; Cong, F.; Liu, Y.G.; Wang, H.M.; Zhang, H.L.; Tian, Z.J.; Tang, Y.D.; Cai, X.H. The Nsp12-coding region of type 2 PRRSV is required for viral subgenomic mRNA synthesis. Emerg. Microbes Infect. 2019, 8, 1501–1510. [Google Scholar] [CrossRef]
- Dong, S.; Liu, L.; Wu, W.; Armstrong, S.D.; Xia, D.; Nan, H.; Hiscox, J.A.; Chen, H. Determination of the interactome of non-structural protein12 from highly pathogenic porcine reproductive and respiratory syndrome virus with host cellular proteins using high throughput proteomics and identification of HSP70 as a cellular factor for virus replication. J. Proteom. 2016, 146, 58–69. [Google Scholar] [CrossRef]
- Li, L.; Zhou, Y.; Jiang, Y.; Gao, F.; Shan, T.; Zhao, K.; Zhang, Y.; Li, L.; Tong, G. Galectin-3 inhibits replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp12 in vitro. Virus Res. 2018, 253, 87–91. [Google Scholar] [CrossRef]
- Yang, L.; Wang, R.; Yang, S.; Ma, Z.; Lin, S.; Nan, Y.; Li, Q.; Tang, Q.; Zhang, Y.J. Karyopherin alpha 6 is required for replication of porcine reproductive and respiratory syndrome virus and zika virus. J. Virol. 2018, 92, e00072-18. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Li, C.; Zhou, L.; Zhang, N.; Wang, X.; Ge, X.; Guo, X.; Yang, H. Porcine reproductive and respiratory syndrome virus counteracts the porcine intrinsic virus restriction factors-IFITM1 and Tetherin in MARC-145 cells. Virus Res. 2014, 191, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Dong, W.; Wang, X.; Zhu, Z.; He, S.; Zhang, H.; Chen, Y.; Liu, X.; Guo, C. Exostosin glycosyltransferase 1 reduces porcine reproductive and respiratory syndrome virus infection through proteasomal degradation of nsp3 and nsp5. J. Biol. Chem. 2022, 298, 101548. [Google Scholar] [CrossRef] [PubMed]
- Wissink, E.H.J.; Kroese, M.V.; Maneschijn-Bonsing, J.G.; Meulenberg, J.J.M.; van Rijn, P.A.; Rijsewijk, F.A.M.; Rottier, P.J.M. Significance of the oligosaccharides of the porcine reproductive and respiratory syndrome virus glycoproteins GP2a and GP5 for infectious virus production. J. Gen. Virol. 2004, 85, 3715–3723. [Google Scholar] [CrossRef] [PubMed]
- Meulenberg, J.J.; Petersen-den Besten, A.; De Kluyver, E.P.; Moormann, R.J.; Schaaper, W.M.; Wensvoort, G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology 1995, 206, 155–163. [Google Scholar] [CrossRef] [Green Version]
- Das, P.B.; Dinh, P.X.; Ansari, I.H.; de Lima, M.; Osorio, F.A.; Pattnaik, A.K. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 2010, 84, 1731–1740. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Chai, Y.; Song, J.; Liu, T.; Chen, P.; Zhou, L.; Ge, X.; Guo, X.; Han, J.; Yang, H. Reprogramming the unfolded protein response for replication by porcine reproductive and respiratory syndrome virus. PLoS Pathog. 2019, 15, e1008169. [Google Scholar] [CrossRef]
- Zhang, M.; Zakhartchouk, A. Porcine reproductive and respiratory syndrome virus envelope (E) protein interacts with tubulin. Vet. Microbiol. 2017, 211, 51–57. [Google Scholar] [CrossRef]
- Pujhari, S.; Zakhartchouk, A.N. Porcine reproductive and respiratory syndrome virus envelope (E) protein interacts with mitochondrial proteins and induces apoptosis. Arch. Virol. 2016, 161, 1821–1830. [Google Scholar] [CrossRef]
- Ke, W.; Fang, L.; Tao, R.; Li, Y.; Jing, H.; Wang, D.; Xiao, S. Porcine reproductive and respiratory syndrome virus e protein degrades porcine cholesterol 25-Hydroxylase via the ubiquitin-proteasome pathway. J. Virol. 2019, 93, e00767-19. [Google Scholar] [CrossRef] [Green Version]
- Gonin, P.; Mardassi, H.; Gagnon, C.A.; Massie, B.; Dea, S. A nonstructural and antigenic glycoprotein is encoded by ORF3 of the IAF-Klop strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 1998, 143, 1927–1940. [Google Scholar] [CrossRef]
- Wieringa, R.; de Vries, A.A.; Raamsman, M.J.; Rottier, P.J. Characterization of two new structural glycoproteins, GP(3) and GP(4), of equine arteritis virus. J. Virol. 2002, 76, 10829–10840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, J.; Zhang, X.; He, B.; Ge, X.; Han, J.; Zhou, L.; Guo, X.; Yang, H. Identification of three site mutations in nonstructural protein 1beta, glycoprotein 3 and glycoprotein 5 that correlate with increased interferon alpha resistance of porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2019, 236, 108395. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Peng, J.; Sun, Y.; Chen, J.; An, T.; Leng, C.; Li, L.; Zhao, H.; Guo, X.; Ge, X.; et al. Unique epitopes recognized by monoclonal antibodies against HP-PRRSV: Deep understanding of antigenic structure and virus-antibody interaction. PLoS ONE 2014, 9, e111633. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Qi, J.; Lu, Y.; Wu, J.; Yoo, D.; Liu, X.; Zhang, X.; Li, J.; Sun, W.; Cong, X.; et al. Evaluation of a DNA vaccine candidate co-expressing GP3 and GP5 of porcine reproductive and respiratory syndrome virus (PRRSV) with interferon alpha/gamma in immediate and long-lasting protection against HP-PRRSV challenge. Virus Genes 2012, 45, 474–487. [Google Scholar] [CrossRef]
- Li, J.; Jiang, P.; Li, Y.; Wang, X.; Cao, J.; Wang, X.; Zeshan, B. HSP70 fused with GP3 and GP5 of porcine reproductive and respiratory syndrome virus enhanced the immune responses and protective efficacy against virulent PRRSV challenge in pigs. Vaccine 2009, 27, 825–832. [Google Scholar] [CrossRef]
- Meulenberg, J.J.; van Nieuwstadt, A.P.; van Essen-Zandbergen, A.; Langeveld, J.P. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J. Virol. 1997, 71, 6061–6067. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.; Lee, C. Proteomic characterization of a novel structural protein ORF5a of porcine reproductive and respiratory syndrome virus. Virus Res. 2012, 169, 255–263. [Google Scholar] [CrossRef]
- Li, L.; Xue, B.; Sun, W.; Gu, G.; Hou, G.; Zhang, L.; Wu, C.; Zhao, Q.; Zhang, Y.; Zhang, G.; et al. Recombinant MYH9 protein C-terminal domain blocks porcine reproductive and respiratory syndrome virus internalization by direct interaction with viral glycoprotein 5. Antivir. Res. 2018, 156, 10–20. [Google Scholar] [CrossRef]
- Xue, B.; Hou, G.; Zhang, G.; Huang, J.; Li, L.; Nan, Y.; Mu, Y.; Wang, L.; Zhang, L.; Han, X.; et al. MYH9 Aggregation Induced by Direct Interaction With PRRSV GP5 Ectodomain Facilitates Viral Internalization by Permissive Cells. Front. Microbiol. 2019, 10, 2313. [Google Scholar] [CrossRef]
- Zhang, M.; Zakhartchouk, A. Characterization of the interactome of the porcine reproductive and respiratory syndrome virus glycoprotein-5. Arch. Virol. 2018, 163, 1595–1605. [Google Scholar] [CrossRef]
- Hicks, J.A.; Yoo, D.; Liu, H.C. Interaction of porcine reproductive and respiratory syndrome virus major envelope proteins GP5 and M with the cellular protein Snapin. Virus Res. 2018, 249, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, J.; Jiang, P.; Li, Y.; Zeshan, B.; Cao, J.; Wang, X. GM-CSF fused with GP3 and GP5 of porcine reproductive and respiratory syndrome virus increased the immune responses and protective efficacy against virulent PRRSV challenge. Virus Res. 2009, 143, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Dea, S.; Gagnon, C.A.; Mardassi, H.; Pirzadeh, B.; Rogan, D. Current knowledge on the structural proteins of porcine reproductive and respiratory syndrome (PRRS) virus: Comparison of the North American and European isolates. Arch. Virol 2000, 145, 659–688. [Google Scholar] [CrossRef] [PubMed]
- Wissink, E.H.; Kroese, M.V.; van Wijk, H.A.; Rijsewijk, F.A.; Meulenberg, J.J.; Rottier, P.J. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J. Virol. 2005, 79, 12495–12506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mardassi, H.; Massie, B.; Dea, S. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 1996, 221, 98–112. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Li, Y.; Dong, H.; Wang, L.; Peng, J.; An, T.; Yang, X.; Tian, Z.; Cai, X. Identification of host cellular proteins that interact with the M protein of a highly pathogenic porcine reproductive and respiratory syndrome virus vaccine strain. Virol. J. 2017, 14, 39. [Google Scholar] [CrossRef] [Green Version]
- Rowland, R.R.; Yoo, D. Nucleolar-cytoplasmic shuttling of PRRSV nucleocapsid protein: A simple case of molecular mimicry or the complex regulation by nuclear import, nucleolar localization and nuclear export signal sequences. Virus Res. 2003, 95, 23–33. [Google Scholar] [CrossRef]
- Forsberg, R. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol. Biol. Evol. 2005, 22, 2131–2134. [Google Scholar] [CrossRef]
- Jourdan, S.S.; Osorio, F.A.; Hiscox, J.A. Biophysical characterisation of the nucleocapsid protein from a highly pathogenic porcine reproductive and respiratory syndrome virus strain. Biochem. Biophys. Res. Commun. 2012, 419, 137–141. [Google Scholar] [CrossRef]
- Song, C.; Lu, R.; Bienzle, D.; Liu, H.C.; Yoo, D. Interaction of the porcine reproductive and respiratory syndrome virus nucleocapsid protein with the inhibitor of MyoD family-a domain-containing protein. Biol. Chem. 2009, 390, 215–223. [Google Scholar] [CrossRef]
- Wang, C.; Zeng, N.; Liu, S.; Miao, Q.; Zhou, L.; Ge, X.; Han, J.; Guo, X.; Yang, H. Interaction of porcine reproductive and respiratory syndrome virus proteins with SUMO-conjugating enzyme reveals the SUMOylation of nucleocapsid protein. PLoS ONE 2017, 12, e0189191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Tian, J.; Nan, H.; Tian, M.; Li, Y.; Xu, X.; Huang, B.; Zhou, E.; Hiscox, J.A.; Chen, H. Porcine Reproductive and Respiratory Syndrome Virus Nucleocapsid Protein Interacts with Nsp9 and Cellular DHX9 To Regulate Viral RNA Synthesis. J. Virol. 2016, 90, 5384–5398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Bai, J.; Fan, B.; Li, Y.; Zhang, Q.; Jiang, P. The Interferon-Induced Mx2 Inhibits Porcine Reproductive and Respiratory Syndrome Virus Replication. J. Interferon Cytokine Res. 2016, 36, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bai, J.; Zhang, L.; Wang, X.; Li, Y.; Jiang, P. Poly(A)-binding protein interacts with the nucleocapsid protein of porcine reproductive and respiratory syndrome virus and participates in viral replication. Antivir. Res. 2012, 96, 315–323. [Google Scholar] [CrossRef]
- Yoo, D.; Wootton, S.K.; Li, G.; Song, C.; Rowland, R.R. Colocalization and interaction of the porcine arterivirus nucleocapsid protein with the small nucleolar RNA-associated protein fibrillarin. J. Virol. 2003, 77, 12173–12183. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Li, L.W.; Jiang, Y.F.; Gao, F.; Zhang, Y.J.; Zhao, W.Y.; Li, G.X.; Yu, L.X.; Zhou, Y.J.; Tong, G.Z. Nucleocapsid protein of porcine reproductive and respiratory syndrome virus antagonizes the antiviral activity of TRIM25 by interfering with TRIM25-mediated RIG-I ubiquitination. Vet. Microbiol. 2019, 233, 140–146. [Google Scholar] [CrossRef]
- Liu, L.; Lear, Z.; Hughes, D.J.; Wu, W.; Zhou, E.M.; Whitehouse, A.; Chen, H.; Hiscox, J.A. Resolution of the cellular proteome of the nucleocapsid protein from a highly pathogenic isolate of porcine reproductive and respiratory syndrome virus identifies PARP-1 as a cellular target whose interaction is critical for virus biology. Vet. Microbiol. 2015, 176, 109–119. [Google Scholar] [CrossRef]
- Sagong, M.; Lee, C. Differential cellular protein expression in continuous porcine alveolar macrophages regulated by the porcine reproductive and respiratory syndrome virus nucleocapsid protein. Virus Res. 2010, 151, 88–96. [Google Scholar] [CrossRef]
- Fang, J.; Wang, H.; Bai, J.; Zhang, Q.; Li, Y.; Liu, F.; Jiang, P. Monkey viperin restricts porcine reproductive and respiratory syndrome virus replication. PLoS ONE 2016, 11, e0156513. [Google Scholar] [CrossRef] [Green Version]
- Jing, H.; Tao, R.; Dong, N.; Cao, S.; Sun, Y.; Ke, W.; Li, Y.; Wang, J.; Zhang, Y.; Huang, H.; et al. Nuclear localization signal in TRIM22 is essential for inhibition of type 2 porcine reproductive and respiratory syndrome virus replication in MARC-145 cells. Virus Genes 2019, 55, 660–672. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Li, L.W.; Zhang, Y.J.; Jiang, Y.F.; Gao, F.; Li, G.X.; Yu, L.X.; Zhao, W.Y.; Shan, T.L.; Zhou, Y.J.; et al. MOV10 inhibits replication of porcine reproductive and respiratory syndrome virus by retaining viral nucleocapsid protein in the cytoplasm of Marc-145cells. Biochem. Biophys. Res. Commun. 2018, 504, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Zhou, Y.; Fang, L.; Ding, Z.; Wang, D.; Ke, W.; Chen, H.; Xiao, S. DExD/H-box helicase 36 signaling via myeloid differentiation primary response gene 88 contributes to NF-kappaB activation to type 2 porcine reproductive and respiratory syndrome virus infection. Front. Immunol. 2017, 8, 1365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, Z.; Bai, J.; Liu, X.; Nauwynck, H.; Wu, J.; Liu, X.; Jiang, P. S100A9 regulates porcine reproductive and respiratory syndrome virus replication by interacting with the viral nucleocapsid protein. Vet. Microbiol. 2019, 239, 108498. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; Wei, R.; Dong, W.; Zhu, Z.; Zhang, X.; Chen, Y.; Liu, X.; Guo, C. CD163(DeltaSRCR5) MARC-145 Cells Resist PRRSV-2 Infection via Inhibiting Virus Uncoating, Which Requires the Interaction of CD163 With Calpain 1. Front. Microbiol. 2019, 10, 3115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhakal, S.; Renukaradhya, G.J. Nanoparticle-based vaccine development and evaluation against viral infections in pigs. Vet. Res. 2019, 50, 90. [Google Scholar] [CrossRef] [Green Version]
- An, T.Q.; Tian, Z.J.; Zhou, Y.J.; Xiao, Y.; Peng, J.M.; Chen, J.; Jiang, Y.F.; Hao, X.F.; Tong, G.Z. Comparative genomic analysis of five pairs of virulent parental/attenuated vaccine strains of PRRSV. Vet. Microbiol. 2011, 149, 104–112. [Google Scholar] [CrossRef]
- Chen, N.; Li, X.; Xiao, Y.; Li, S.; Zhu, J. Characterization of four types of MLV-derived porcine reproductive and respiratory syndrome viruses isolated in unvaccinated pigs from 2016 to 2020. Res. Vet. Sci. 2021, 134, 102–111. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.; Duan, H.; Zhang, A.; Liang, C.; Gao, J.; Zhang, C.; Huang, B.; Li, Q.; Li, N.; et al. An intracellularly expressed Nsp9-specific nanobody in MARC-145 cells inhibits porcine reproductive and respiratory syndrome virus replication. Vet. Microbiol. 2015, 181, 252–260. [Google Scholar] [CrossRef]
- Liu, H.; Liang, C.; Duan, H.; Zhang, X.; Wang, X.; Xiao, S.; Zhou, E.M. Intracellularly expressed nanobodies against non-structural protein 4 of porcine reproductive and respiratory syndrome virus inhibit virus replication. Biotechnol. Lett. 2016, 38, 1081–1088. [Google Scholar] [CrossRef]
- Tang, C.; Deng, Z.; Li, X.; Yang, M.; Tian, Z.; Chen, Z.; Wang, G.; Wu, W.; Feng, W.H.; Zhang, G.; et al. Helicase of type 2 porcine reproductive and respiratory syndrome virus strain HV reveals a unique structure. Viruses 2020, 12, 215. [Google Scholar] [CrossRef] [Green Version]
- Xue, F.; Sun, Y.; Yan, L.; Zhao, C.; Chen, J.; Bartlam, M.; Li, X.; Lou, Z.; Rao, Z. The crystal structure of porcine reproductive and respiratory syndrome virus nonstructural protein Nsp1beta reveals a novel metal-dependent nuclease. J. Virol. 2010, 84, 6461–6471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Z.; Lehmann, K.C.; Li, X.; Feng, C.; Wang, G.; Zhang, Q.; Qi, X.; Yu, L.; Zhang, X.; Feng, W.; et al. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014, 42, 3464–3477. [Google Scholar] [CrossRef]
- Zhang, M.; Li, X.; Deng, Z.; Chen, Z.; Liu, Y.; Gao, Y.; Wu, W.; Chen, Z. Structural biology of the arterivirus nsp11 endoribonucleases. J. Virol. 2017, 91, e01309-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zheng, Z.; Zhou, P.; Zhang, B.; Shi, Z.; Hu, Q.; Wang, H. The cysteine protease domain of porcine reproductive and respiratory syndrome virus non-structural protein 2 antagonizes interferon regulatory factor 3 activation. J. Gen. Virol. 2010, 91, 2947–2958. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shi, X.; Zhang, X.; Wang, A.; Wang, L.; Chen, J.; Deng, R.; Zhang, G. The Endoribonuclease Activity Essential for the Nonstructural Protein 11 of Porcine Reproductive and Respiratory Syndrome Virus to Inhibit NLRP3 Inflammasome-Mediated IL-1beta Induction. DNA Cell Biol. 2015, 34, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Nan, Y.; Xiao, S.; Zhao, Q.; Zhou, E.M. Antiviral Strategies against PRRSV Infection. Trends Microbiol. 2017, 25, 968–979. [Google Scholar] [CrossRef]
- Whitworth, K.M.; Rowland, R.R.; Ewen, C.L.; Trible, B.R.; Kerrigan, M.A.; Cino-Ozuna, A.G.; Samuel, M.S.; Lightner, J.E.; McLaren, D.G.; Mileham, A.J.; et al. Gene-edited pigs are protected from porcine reproductive and respiratory syndrome virus. Nat. Biotechnol. 2016, 34, 20–22. [Google Scholar] [CrossRef]
- Burkard, C.; Opriessnig, T.; Mileham, A.J.; Stadejek, T.; Ait-Ali, T.; Lillico, S.G.; Whitelaw, C.B.A.; Archibald, A.L. Pigs lacking the scavenger receptor cysteine-rich domain 5 of CD163 are resistant to porcine reproductive and respiratory syndrome virus 1 infection. J. Virol. 2018, 92, e00415-18. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Zhang, J.; Zhang, X.; Shi, J.; Pan, Y.; Zhou, R.; Li, G.; Li, Z.; Cai, G.; Wu, Z. CD163 knockout pigs are fully resistant to highly pathogenic porcine reproductive and respiratory syndrome virus. Antivir. Res. 2018, 151, 63–70. [Google Scholar] [CrossRef]
- Prather, R.S.; Wells, K.D.; Whitworth, K.M.; Kerrigan, M.A.; Samuel, M.S.; Mileham, A.; Popescu, L.N.; Rowland, R.R.R. Knockout of maternal CD163 protects fetuses from infection with porcine reproductive and respiratory syndrome virus (PRRSV). Sci. Rep. 2017, 7, 13371. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Jiang, L.; Qiao, S.; Zhi, Y.; Chen, X.X.; Yang, Y.; Huang, X.; Huang, M.; Li, R.; Zhang, G.P. The crystal structure of the fifth scavenger receptor cysteine-rich domain of porcine CD163 Reveals an important residue involved in porcine reproductive and respiratory syndrome virus infection. J. Virol. 2017, 91, e01897-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Sha, H.; Qin, L.; Wang, N.; Kong, W.; Huang, L.; Zhao, M. Research Progress in Porcine Reproductive and Respiratory Syndrome Virus–Host Protein Interactions. Animals 2022, 12, 1381. https://doi.org/10.3390/ani12111381
Zhang H, Sha H, Qin L, Wang N, Kong W, Huang L, Zhao M. Research Progress in Porcine Reproductive and Respiratory Syndrome Virus–Host Protein Interactions. Animals. 2022; 12(11):1381. https://doi.org/10.3390/ani12111381
Chicago/Turabian StyleZhang, Hang, Huiyang Sha, Limei Qin, Nina Wang, Weili Kong, Liangzong Huang, and Mengmeng Zhao. 2022. "Research Progress in Porcine Reproductive and Respiratory Syndrome Virus–Host Protein Interactions" Animals 12, no. 11: 1381. https://doi.org/10.3390/ani12111381





