Differential Responses of Digesta- and Mucosa-Associated Jejunal Microbiota of Hu Sheep to Pelleted and Non-Pelleted High-Grain Diets
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Animals
2.2. Sample Collection
2.3. Genomic DNA Isolation
2.4. Illumina Sequencing and Data Analysis
2.5. The Prediction of Microbial Functional Genes
2.6. Statistical Analysis
3. Results
3.1. Overview of Jejunal Digesta- and Mucosa-Associated Microbiota
3.2. Diversity and Richness of Jejunal Microbiota
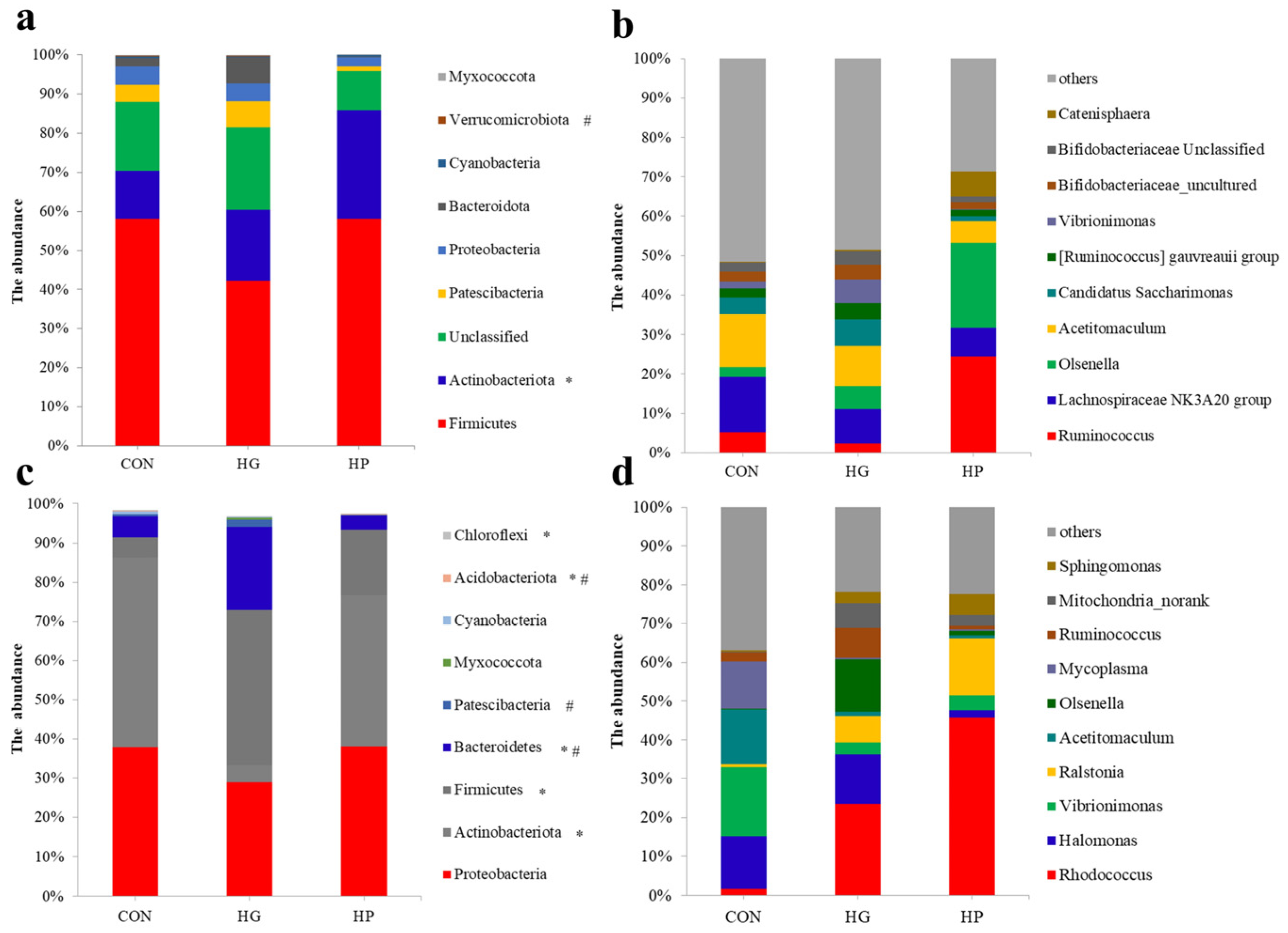
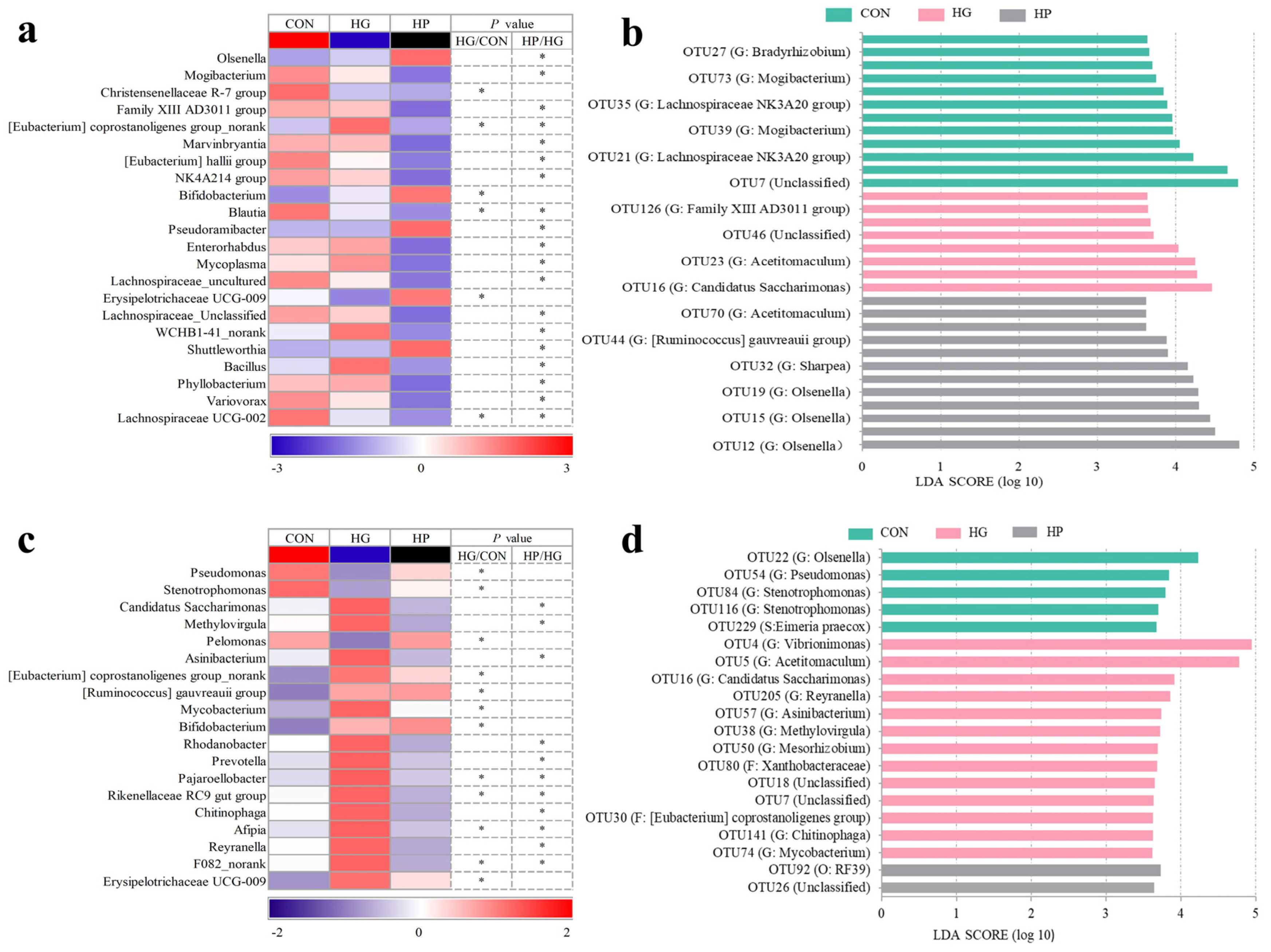
3.3. Alterations in the Microbial Composition of the Jejunal Digesta
3.4. Alterations in the Microbial Composition of the Jejunal Mucosa
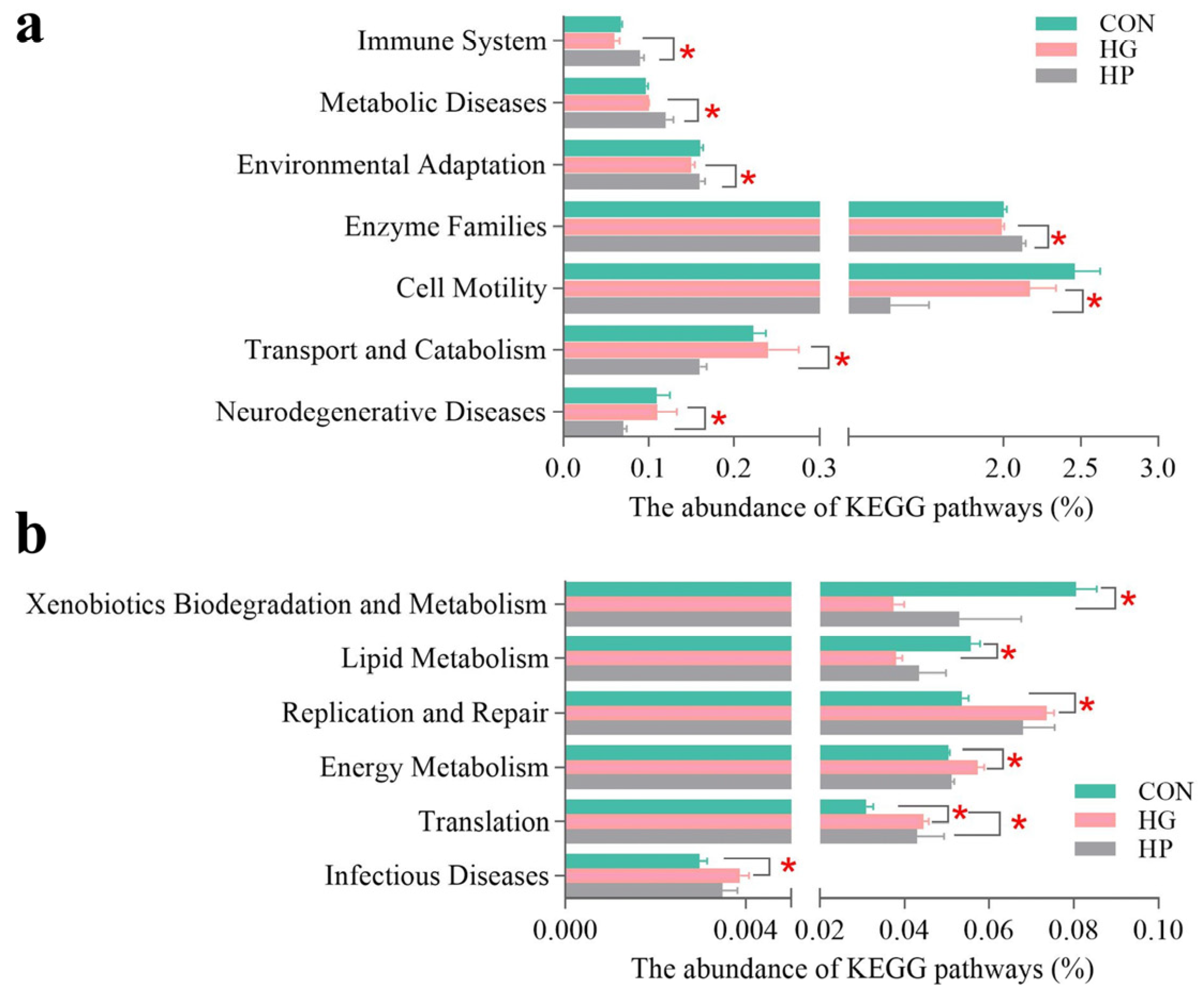
3.5. Changes in the Microbial Functions of Jejunal Microbiota
4. Discussion
4.1. Dietary Treatments Led to Differential Response Changes in Microbial Richness and Diversity in Different Ecological Niches
4.2. Changes in the Microbial Structure of Jejunal Digesta after Different Dietary Treatments
4.3. Changes of the Microbial Community in Jejunal Mucosa-Associated Microbiota after Various Dietary Treatments
4.4. Alterations in Jejunal Microbiota Resulted in Distinct Microbial Functions after Different Diets
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Krause, K.; Oetzel, G. Inducing subacute ruminal acidosis in lactating dairy cows. J. Dairy Sci. 2005, 88, 3633–3639. [Google Scholar] [CrossRef]
- Gao, X.; Oba, M. Relationship of severity of subacute ruminal acidosis to rumen fermentation, chewing activities, sorting behavior, and milk production in lactating dairy cows fed a high-grain diet. J. Dairy Sci. 2014, 97, 3006–3016. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.-H.; Bian, G.-R.; Zhu, W.-Y.; Mao, S.-Y. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015, 6, 167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a high concentration diet induces unhealthy alterations in the composition and metabolism of ruminal microbiota and host response in a goat model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, R.; Jin, W.; Feng, P.; Liu, J.; Mao, S. High-grain diet feeding altered the composition and functions of the rumen bacterial community and caused the damage to the laminar tissues of goats. Animal 2018, 12, 2511–2520. [Google Scholar] [CrossRef]
- Metzler-Zebeli, B.U.; Schmitz-Esser, S.; Klevenhusen, F.; Podstatzky-Lichtenstein, L.; Wagner, M.; Zebeli, Q. Grain-rich diets differently alter ruminal and colonic abundance of microbial populations and lipopolysaccharide in goats. Anaerobe 2013, 20, 65–73. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Liu, J.; Zhu, W.; Mao, S. A high grain diet dynamically shifted the composition of mucosa-associated microbiota and induced mucosal injuries in the colon of sheep. Front. Microbiol. 2017, 8, 2080. [Google Scholar] [CrossRef] [Green Version]
- Ye, H.; Liu, J.; Feng, P.; Zhu, W.; Mao, S. Grain-rich diets altered the colonic fermentation and mucosa-associated bacterial communities and induced mucosal injuries in goats. Sci. Rep. 2016, 6, 20329. [Google Scholar]
- Bonfante, E.; Palmonari, A.; Mammi, L.; Canestrari, G.; Fustini, M.; Formigoni, A. Effects of a completely pelleted diet on growth performance in Holstein heifers. J. Dairy Sci. 2016, 99, 9724–9731. [Google Scholar] [CrossRef] [Green Version]
- Owens, F.N.; Secrist, D.S.; Hill, W.J.; Gill, D.R. The effect of grain source and grain processing on performance of feedlot cattle: A review. J. Anim. Sci. 1997, 75, 868–879. [Google Scholar] [CrossRef] [Green Version]
- Blanco, C.; Giráldez, F.J.; Prieto, N.; Benavides, J.; Wattegedera, S.; Morán, L.; Andrés, S.; Bodas, R. Total mixed ration pellets for light fattening lambs: Effects on animal health. Animal 2015, 9, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Fang, Y.; Zhou, D.; Sun, X.; Zhou, C.; He, Y. Pelleted total mixed ration improves growth performance of fattening lambs. Anim. Feed Sci. Technol. 2018, 242, 127–134. [Google Scholar] [CrossRef]
- Li, B.; Sun, X.; Huo, Q.; Zhang, G.; Wu, T.; You, P.; He, Y.; Tian, W.; Li, R.; Li, C. Pelleting of a total mixed ration affects growth performance of fattening lambs. Front. Vet. Sci. 2021, 8, 629016. [Google Scholar] [CrossRef] [PubMed]
- Trabi, E.B.; Seddik, H.-e.; Xie, F.; Lin, L.; Mao, S. Comparison of the rumen bacterial community, rumen fermentation and growth performance of fattening lambs fed low-grain, pelleted or non-pelleted high grain total mixed ration. Anim. Feed Sci. Technol. 2019, 253, 1–12. [Google Scholar]
- Lin, L.; Trabi, E.B.; Xie, F.; Mao, S. Comparison of the fermentation and bacterial community in the colon of Hu sheep fed a low-grain, non-pelleted, or pelleted high-grain diet. Appl. Microbiol. Biotechnol. 2021, 105, 2071–2080. [Google Scholar] [CrossRef]
- Myer, P.; Wells, J.; Smith, T.; Kuehn, L.; Freetly, H. Microbial community profiles of the jejunum from steers differing in feed efficiency. J. Anim. Sci. 2016, 94, 327–338. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, J.; Zhou, C.; Tan, Z.; Jiao, J. Spatial and temporal organization of jejunal microbiota in goats during animal development process. J. Appl. Microbiol. 2021, 131, 68–79. [Google Scholar] [CrossRef]
- Wen, K.; Zhao, M.; Liu, L.; Khogali, M.K.; Geng, T.; Wang, H.; Gong, D. Thiamine modulates intestinal morphological structure and microbiota under subacute ruminal acidosis induced by a high-concentrate diet in Saanen goats. Animal 2021, 15, 100370. [Google Scholar] [CrossRef]
- Yuan, C.; Graham, M.; Staley, C.; Subramanian, S. Mucosal microbiota and metabolome along the intestinal tract reveal a location-specific relationship. Msystems 2020, 5, e00055-20. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Griebel, P.J.; Guan, L.L. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl. Environ. Microbiol. 2014, 80, 2021–2028. [Google Scholar] [CrossRef] [Green Version]
- Trabi, E.B.; Seddik, H.; Xie, F.; Wang, X.; Liu, J.; Mao, S. Effect of pelleted high-grain total mixed ration on rumen morphology, epithelium-associated microbiota and gene expression of proinflammatory cytokines and tight junction proteins in Hu sheep. Anim. Feed Sci. Technol. 2020, 263, 114453. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Consortium, H.M.P. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Tuddenham, S.; Sears, C.L. The intestinal microbiome and health. Curr. Opin. Infect. Dis. 2015, 28, 464–470. [Google Scholar] [CrossRef] [Green Version]
- Fernando, S.C.; Purvis, H.; Najar, F.; Sukharnikov, L.; Krehbiel, C.; Nagaraja, T.; Roe, B.; Desilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microbiol. 2010, 76, 7482–7490. [Google Scholar] [CrossRef] [Green Version]
- Plaizier, J.C.; Li, S.; Tun, H.M.; Khafipour, E. Nutritional models of experimentally-induced subacute ruminal acidosis (SARA) differ in their impact on rumen and hindgut bacterial communities in dairy cows. Front. Microbiol. 2017, 7, 2128. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xu, T.; Zhu, W.; Mao, S. High-grain feeding alters caecal bacterial microbiota composition and fermentation and results in caecal mucosal injury in goats. Br. J. Nutr. 2014, 112, 416–427. [Google Scholar] [CrossRef] [Green Version]
- Plaizier, J.C.; Danscher, A.-M.; Azevedo, P.A.; Derakhshani, H.; Andersen, P.H.; Khafipour, E. A Grain-Based SARA Challenge Affects the Composition of Epimural and Mucosa-Associated Bacterial Communities throughout the Digestive Tract of Dairy Cows. Animals 2021, 11, 1658. [Google Scholar] [CrossRef] [PubMed]
- Gharechahi, J.; Vahidi, M.F.; Bahram, M.; Han, J.-L.; Ding, X.-Z.; Salekdeh, G.H. Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 2021, 15, 1108–1120. [Google Scholar] [CrossRef] [PubMed]
- Bunesova, V.; Vlkova, E.; Rada, V.; Killer, J.; Musilova, S. Bifidobacteria from the gastrointestinal tract of animals: Differences and similarities. Benef. Microbes 2014, 5, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Pokusaeva, K.; Fitzgerald, G.F.; van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rainey, F.A. Acetitomaculum. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2015; pp. 1–4. [Google Scholar]
- Greening, R.; Leedle, J. Enrichment and isolation of Acetitomaculum ruminis, gen. nov., sp. nov.: Acetogenic bacteria from the bovine rumen. Arch. Microbiol. 1989, 151, 399–406. [Google Scholar] [CrossRef]
- Tong, J.; Zhang, H.; Yang, D.; Zhang, Y.; Xiong, B.; Jiang, L. Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS ONE 2018, 13, e0198225. [Google Scholar]
- Guo, J.; Mu, R.; Li, S.; Zhang, N.; Fu, Y.; Hu, X. Characterization of the Bacterial Community of Rumen in Dairy Cows with Laminitis. Genes 2021, 12, 1996. [Google Scholar] [CrossRef]
- Fan, Q.; Wanapat, M.; Hou, F. Rumen bacteria influence milk protein yield of yak grazing on the Qinghai-Tibet plateau. Anim. Biosci. 2021, 34, 1466–1478. [Google Scholar] [CrossRef]
- Mancabelli, L.; Milani, C.; Lugli, G.A.; Turroni, F.; Cocconi, D.; van Sinderen, D.; Ventura, M. Identification of universal gut microbial biomarkers of common human intestinal diseases by meta-analysis. FEMS Microbiol. Ecol. 2017, 93, fix153. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A new functional genus with potential probiotic properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef] [PubMed]
- Plaizier, J.; Azevedo, P.; Schurmann, B.; Górka, P.; Penner, G.; Khafipour, E. The duration of increased grain feeding affects the microbiota throughout the digestive tract of yearling Holstein steers. Microorganisms 2020, 8, 1854. [Google Scholar] [CrossRef] [PubMed]
- Gaowa, N.; Li, W.; Gelsinger, S.; Murphy, B.; Li, S. Analysis of Host Jejunum Transcriptome and Associated Microbial Community Structure Variation in Young Calves with Feed-Induced Acidosis. Metabolites 2021, 11, 414. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-W.; Jia, H.-J.; Zhang, H.-J.; Wang, J.; Lv, H.-Y.; Wu, S.-G.; Qi, G.-H. Supplemental plant extracts from Flos lonicerae in combination with Baikal skullcap attenuate intestinal disruption and modulate gut microbiota in laying hens challenged by Salmonella pullorum. Front. Microbiol. 2019, 10, 1681. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.; Tian, P.; Luo, Y.; Tian, J.; Hua, C.; Geng, Y.; Cong, R.; Ni, Y.; Zhao, R. Microbiome-metabolome responses to a high-grain diet associated with the hind-gut health of goats. Front. Microbiol. 2017, 8, 1764. [Google Scholar] [CrossRef]
- O’Hara, E.; Kelly, A.; McCabe, M.S.; Kenny, D.A.; Guan, L.L.; Waters, S.M. Effect of a butyrate-fortified milk replacer on gastrointestinal microbiota and products of fermentation in artificially reared dairy calves at weaning. Sci. Rep. 2018, 8, 14901. [Google Scholar] [CrossRef]
- Kong, F.; Gao, Y.; Tang, M.; Fu, T.; Diao, Q.; Bi, Y.; Tu, Y. Effects of dietary rumen–protected Lys levels on rumen fermentation and bacterial community composition in Holstein heifers. Appl. Microbiol. Biotechnol. 2020, 104, 6623–6634. [Google Scholar] [CrossRef]
- Chen, W.; Liu, F.; Ling, Z.; Tong, X.; Xiang, C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS ONE 2012, 7, e39743. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Q.; Sun, S.; Huang, B.; Zhang, Y.; Xu, Y.; Zhang, S.; Xiang, H. Probiotics-fermented Massa Medicata Fermentata ameliorates weaning stress in piglets related to improving intestinal homeostasis. Appl. Microbiol. Biotechnol. 2018, 102, 10713–10727. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, L.; Hu, M.; Kim, J.J.; Lin, R.; Xu, J.; Fan, L.; Qi, Y.; Wang, L.; Liu, W. Dietary type 2 resistant starch improves systemic inflammation and intestinal permeability by modulating microbiota and metabolites in aged mice on high-fat diet. Aging 2020, 12, 9173–9187. [Google Scholar] [CrossRef]
- Carroll, I.M.; Chang, Y.-H.; Park, J.; Sartor, R.B.; Ringel, Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010, 2, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinsen, F.-A.; Knecht, H.; Neulinger, S.C.; Schmitz, R.A.; Knecht, C.; Kühbacher, T.; Rosenstiel, P.C.; Schreiber, S.; Friedrichs, A.K.; Ott, S.J. Dynamic changes of the luminal and mucosa-associated gut microbiota during and after antibiotic therapy with paromomycin. Gut Microbes 2015, 6, 243–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Q.; Gao, C.; Aziz ur Rahman, M.; Cao, B.; Su, H. Digestive ability, physiological characteristics, and rumen bacterial community of holstein finishing steers in response to three nutrient density diets as fattening phases advanced. Microorganisms 2020, 8, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holman, D.B.; Yang, W.; Alexander, T.W. Antibiotic treatment in feedlot cattle: A longitudinal study of the effect of oxytetracycline and tulathromycin on the fecal and nasopharyngeal microbiota. Microbiome 2019, 7, 86. [Google Scholar] [CrossRef] [Green Version]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef] [Green Version]
- Davis, W.C.; Madsen-Bouterse, S.A. Crohn’s disease and Mycobacterium avium subsp. paratuberculosis: The need for a study is long overdue. Vet. Immunol. Immunopathol. 2012, 145, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Ntaikou, I.; Gavala, H.N.; Kornaros, M.; Lyberatos, G. Hydrogen production from sugars and sweet sorghum biomass using Ruminococcus albus. Int. J. Hydrogen Energy 2008, 33, 1153–1163. [Google Scholar] [CrossRef]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.L.Q.; Anani, H.; Trinh, H.T.; Pham, T.P.T.; Ho, V.M.; Bui, N.H.L.; Nguyen, N.H.; Raoult, D.; Trinh, T.T.; Fournier, P.E. Chitinophaga vietnamensis sp. nov., a multi-drug resistant bacterium infecting humans. Int. J. Syst. Evol. Microbiol. 2020, 70, 1758–1768. [Google Scholar] [CrossRef]
- Cortés, A.; Wills, J.; Su, X.; Hewitt, R.E.; Robertson, J.; Scotti, R.; Price, D.R.; Bartley, Y.; McNeilly, T.N.; Krause, L. Infection with the sheep gastrointestinal nematode Teladorsagia circumcincta increases luminal pathobionts. Microbiome 2020, 8, 60. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, X.; Wang, M.; Zhou, C.; Yan, Q.; Tan, Z. Linkages between epithelial microbiota and host transcriptome in the ileum during high-grain challenges: Implications for gut homeostasis in goats. J. Agric. Food Chem. 2018, 67, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Dashty, M. A quick look at biochemistry: Carbohydrate metabolism. Clin. Biochem. 2013, 46, 1339–1352. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.F. Oxidative phosphorylation: Regulation and role in cellular and tissue metabolism. J. Physiol. 2017, 595, 7023–7038. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Khafipour, E.; Krause, D.; Kroeker, A.; Rodriguez-Lecompte, J.; Gozho, G.; Plaizier, J. Effects of subacute ruminal acidosis challenges on fermentation and endotoxins in the rumen and hindgut of dairy cows. J. Dairy Sci. 2012, 95, 294–303. [Google Scholar] [CrossRef]
- Nagaraja, T.; Bartley, E.; Fina, L.; Anthony, H. Relationship of rumen gram-negative bacteria and free endotoxin to lactic acidosis in cattle. J. Anim. Sci. 1978, 47, 1329–1337. [Google Scholar] [CrossRef] [PubMed]
- Siebold, C.; Flükiger, K.; Beutler, R.; Erni, B. Carbohydrate transporters of the bacterial phosphoenolpyruvate: Sugar phosphotransferase system (PTS). FEBS Lett. 2001, 504, 104–111. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhong, Z.; Zhang, Y.; Li, X.; Li, L.; Zhang, R.; Zhang, S. Differential Responses of Digesta- and Mucosa-Associated Jejunal Microbiota of Hu Sheep to Pelleted and Non-Pelleted High-Grain Diets. Animals 2022, 12, 1695. https://doi.org/10.3390/ani12131695
Zhong Z, Zhang Y, Li X, Li L, Zhang R, Zhang S. Differential Responses of Digesta- and Mucosa-Associated Jejunal Microbiota of Hu Sheep to Pelleted and Non-Pelleted High-Grain Diets. Animals. 2022; 12(13):1695. https://doi.org/10.3390/ani12131695
Chicago/Turabian StyleZhong, Zhiqiang, Yuning Zhang, Xiaotong Li, Lingyun Li, Ruiyang Zhang, and Shuyi Zhang. 2022. "Differential Responses of Digesta- and Mucosa-Associated Jejunal Microbiota of Hu Sheep to Pelleted and Non-Pelleted High-Grain Diets" Animals 12, no. 13: 1695. https://doi.org/10.3390/ani12131695
APA StyleZhong, Z., Zhang, Y., Li, X., Li, L., Zhang, R., & Zhang, S. (2022). Differential Responses of Digesta- and Mucosa-Associated Jejunal Microbiota of Hu Sheep to Pelleted and Non-Pelleted High-Grain Diets. Animals, 12(13), 1695. https://doi.org/10.3390/ani12131695



