1. Introduction
Cashmere fibers are non-medullated and produced by the secondary follicles in cashmere goat skin. They are used to manufacture luxury textile products due to their softness, lightness, and warmth. The yield of cashmere fiber, the mean fiber diameter (MFD), and fiber length are important traits as they determine the economic return for cashmere production [
1]. These are moderate to highly heritable traits [
2]. As the quantity and quality of cashmere fibers are affected by environmental, genetic, and nutritional influences, the identification of genes that affect cashmere fiber traits provides one basis for fiber improvement.
The fibers from cashmere goats contain keratin (K) proteins and keratin-associated proteins (KAPs), with the Ks assembled into keratin intermediate filaments (KIFs), while KAPs form a milieu that cross-links the KIFs [
3]. Consequently, the Ks and KAPs have a role in defining the properties of fibers.
The KAPs are proteins that range from 10–30 kDa in size and usually have either a high content of the amino acid cysteine, or glycine and tyrosine [
3,
4]. They have been categorized into three groups: high-glycine/tyrosine KAPs (35–60 mol % glycine and tyrosine), high-sulfur KAPs (≤30 mol % cysteine), and ultra-high-sulfur KAPS (>30 mol % cysteine) [
5].
The KAPs are encoded by small intron-less genes called
KRTAPs [
5]. While 88 functional
KRTAPs from 25 families have been identified in humans, and 30
KRTAPs comprised of 18 families have been reported in sheep, only 18
KRTAPs and 12 families having been identified in goats [
6]. Of the 18 caprine
KRTAPs identified, variation in 10 of these genes has been reported to be associated with selected cashmere fiber traits, including
KRTAP1-3 [
7],
KRTAP8-2 [
8],
KRTAP9-2 [
9],
KRTAP13-1 [
10],
KRTAP15-1 [
11],
KRTAP20-1 [
12],
KRTAP20-2 [
1],
KRTAP24-1 [
13],
KRTAP27-1 [
14], and
KRTAP28-1 [
15].
Previous studies have identified HGT-
KRTAPs that are present in sheep and goats but are absent in humans [
8,
16,
17,
18], with this suggesting a difference in the complexity of HGT-KAPs between wool/cashmere fibers and human hair. Of all the HGT-
KRTAPs identified for sheep, the highest level of heterogeneity is observed for the KAP6 family, for which five members are present in sheep compared to three in humans [
17,
19].
No
KRTAPs in the KAP6 family have been identified in goats. Despite there being two goat
KRTAP sequences (EU145019 and AY316158; [
20,
21]) reported to be “alleles of caprine
KRTAP6-2”, these two sequences do not share high similarity to any
KRTAP6-n sequences from sheep or humans [
6]. This suggests that the reported sequences may not be from the caprine
KRTAP6-2, and that caprine
KRTAP6-2 remains to be identified.
In this research, we aimed to find the KAP gene KRTAP6-2 in cashmere producing Longdong goats. We used a polymerase chain reaction-single strand conformation polymorphism (PCR-SSCP) analysis to distinguish nucleotide sequence differences in the gene, and to investigate whether these differences were predictive of variation in selected important cashmere fiber traits.
2. Materials and Methods
2.1. Description of the Longdong Casgmere Goats Investigated, and Sample and Data Collection
Longdong cashmere goats (
n = 230 in total) were studied. These were raised in the Gansu Province of China by the Yusheng Cashmere Goat Breeding Company. When the goats were approximately one year old, their fleece was combed to retrieve the cashmere fiber, as is the traditional practice. The weight of fiber retrieved for each goat was recorded. A small sample of fiber was specifically collected from each goat’s mid-side region to enable the determination of the mean fiber diameter (MFD) and fiber length using the Optical Fiber Length and Diameter Analyzer (OFDA4000; EPCO, Shanghai, China). At the time of fiber collection, a small sample of blood was retrieved from the ear of each goat and absorbed onto separate FTA
TM cards (Whatman BioScience, Middlesex, UK). The samples were air-dried and stored in the dark at room temperature until required for further analysis. Genomic DNA that binds to the FTA
TM was prepared for PCR amplification using an approach defined by Zhou et al. [
22]. Briefly, a 1.2 mm disk was excised from the blood sample on the card and put in a 0.7-mL tube containing 200 µL of 20 mM NaOH. These samples were incubated for 20 to 30 min at 60 °C. The liquid was then aspirated, and the disk equilibrated in 200 µL of TE
−1 buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0). The buffer was then removed, and the disks allowed to air dry.
2.2. PCR-SSCP Analysis of Caprine KRTAP6-2
To identify caprine KRTAP6-2, an ovine gene sequence (accession number KT725827) was used to search the caprine ASM170441v1 genome assembly. The sequence that shared the most similarity to ovine KRTAP6-2 was expected to be caprine KRTAP6-2. Based on this caprine sequence, PCR primers (5′-GAGAAATGTCCACACTCAAGT-3′ and 5′-GAGGGCATTAAAAGGCACGT-3′; synthesized by the Takara Biotechnology Co., Ltd., Dalian, China) were designed to amplify a 430-bp portion of the caprine DNA that encompassed the entire coding region of what was presumed to be the caprine KRTAP6-2.
Sequence amplification with PCR was undertaken in a 20-µL reaction comprising the genomic DNA on the 1.2-mm punch of blood, 0.25 µM of each primer, 150 µM of the four dNTPs (Takara, Dalian, China), 2.5 mM Mg2+, 0.5U of Taq DNA polymerase (Takara, Dalian, China), and 1× PCR reaction buffer that was supplied with the polymerase enzyme. The thermal cycling procedure included incubation for 2 min at 94 °C, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. A final extension of 5 min at 72 °C was used to ‘polish’ any incomplete amplicons. Thermal cycling was carried out in Bio-Rad S1000 thermal cyclers (Bio-Rad, Hercules, CA, USA).
Upon completion of the amplifications, the amplicons were examined using a SSCP method. For these analyses, a 1-µL aliquot of the amplicon was combined with 7 µL of loading dye (98% formamide, 10 mM EDTA, 0.025% bromophenol blue and 0.025% xylene-cyanol). These samples were incubated at 95 °C for 10 min to denature the DNA to a single-strand state, then quickly cooled on wet ice. They were immediately loaded onto 16 × 18 cm, 12% acrylamide: bisacrylamide (37.5:1) (Bio-Rad) gels that contained 0.2%
v/
v glycerol. Electrophoresis was performed in 0.5× TBE buffer at 230 volts and 18 °C for 21 h. A method described by Byun et al. [
23] was used to stain the gels and reveal band patterns.
2.3. DNA Sequencing and Sequence Analyses
Following the PCR-SSCP analysis, amplicons that were observed to be homozygous were subjected to direct DNA sequence using the original PCR primers. However, for amplicons that were only found in a heterozygous form, the DNA templates for subsequent sequencing were produced using an approach that has been described by Gong et al. [
24]. All the DNA sequencing was conducted at the Beijing Genomics Institute (Beijing, China).
The online DNA sequence analysis tool Open Reading Frame Finder (
https://www.ncbi.nlm.nih.gov/orffinder/) was used to find any open reading frames in the DNA sequences that were produced. DNAMAN (version 5.2.10 Lynnon BioSoft, Vaudreuil, Canada) was then used to align and compare DNA sequences, and to construct phylogenetic trees. The BLAST algorithm (
http://blast.ncbi.nlm.nih.gov/; accessed on 21 May 2022) was used to search the NCBI GenBank databases for sequences homologous to those obtained from the amplicons.
2.4. Statistical Analyses
All data were analyzed using SPSS v24.0 (IBM, Armonk, NY, USA). General linear mixed models (GLMMs) were used to evaluate associations between the presence or absence of individual variants of caprine KRTAP6-2 and variation in the crimped fiber length, cashmere yield and MFD. Gender and sire were revealed in ANOVAs to affect all the fiber traits (p < 0.05), so they were fitted as fixed and random factors, respectively. Birth rank did not affect fiber traits (p > 0.05), so it was not included as a factor in the models.
3. Results
3.1. Identification of Caprine KRTAP6-2
A BLAST search of the Caprine Genome Assembly NC_030808.1 using an ovine KRTAP6-2 sequence (KT725827.1) revealed a highly similar region (identity 97%, E value 0.0) on chromosome 1. A 252-bp open reading frame (ORF) was found within this region at the location NC_030808.1: nt3536200-nt3536451.
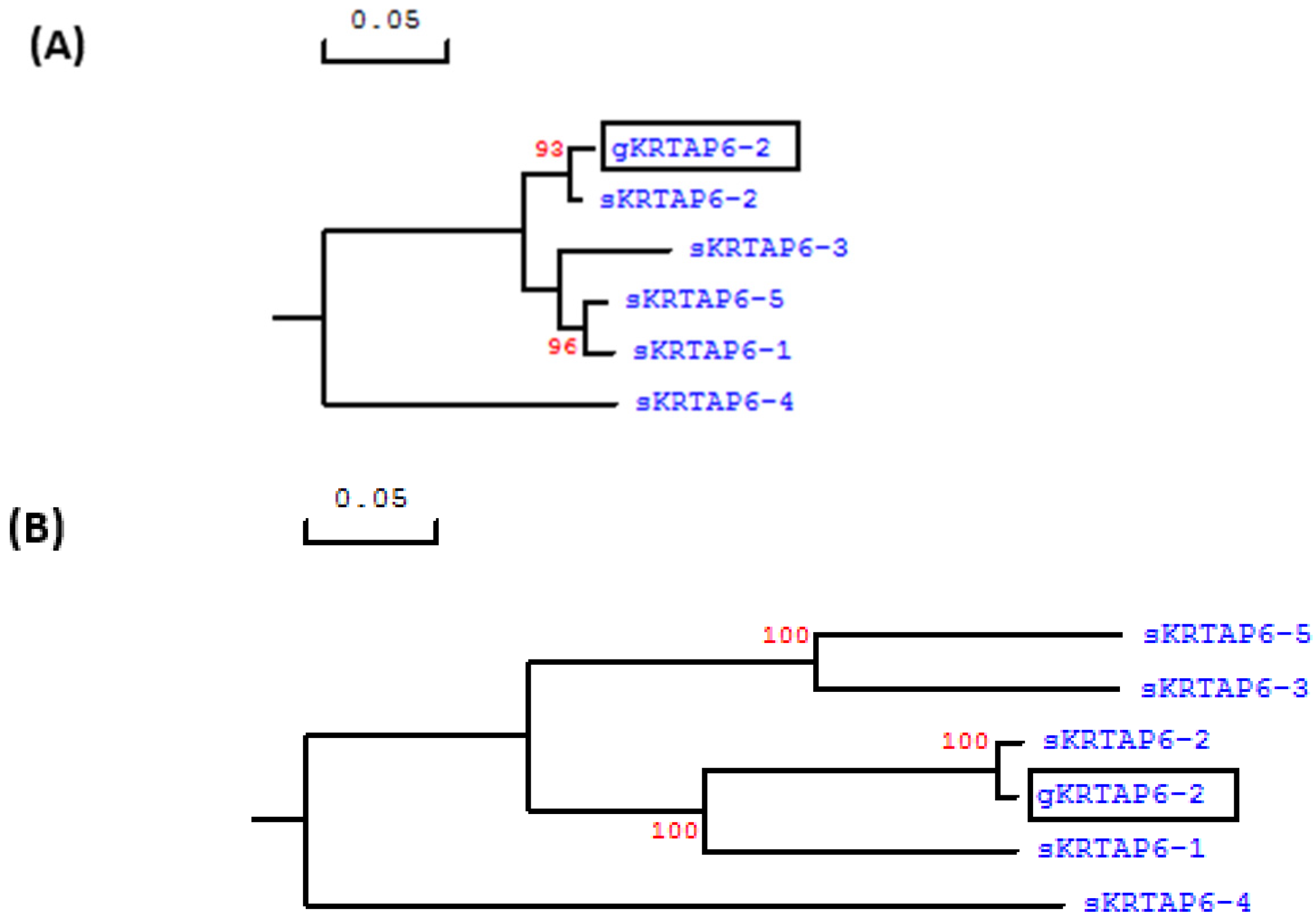
As the
KRTAP6-n in sheep tend to share an elevated level of sequence similarity in the coding region, to confirm that this 252-bp ORF represented caprine
KRTAP6-2, the 300-bp sequences upstream and downstream of this ORF in the Caprine Genome Assembly were also analyzed. A phylogenetic assessment confirmed that the upstream and downstream sequences of the ORF were more closely related to ovine
KRTAP6-2 than any other
KRTAP6-n sequences from sheep (
Figure 1). The location of this caprine ORF was found to be approximately 50 kb downstream of the
KRTAP20-2 sequence and approximately 89 kb upstream of
KRTAP20-1.
3.2. Identification of Nucleotide Sequence Variation in Caprine KRTAP6-2
In the 230 goats studied, different PCR-SSCP banding patterns were observed and these were resolved to suggest that there were five unique nucleotide sequences present in both homozygous and heterozygous genotypes (
Figure 2). DNA sequencing of the PCR amplicons subsequently revealed five nucleotide sequences (named
A to
E; GenBank accession numbers OP157192-OP157196). All these sequences were different, but shared high sequence similarities (over 98%) to the goat
KRTAP6-2 sequence identified in the goat genome assembly sequence NC_030808.1. This suggests that these sequences were derived from the same
KRTAP6-2 locus and that the differences in the sequences may represent variation in the gene.
Among the five caprine
KRTAP6-2 variants identified, there were 11 SNPs, a three-nucleotide sequence variation and an insertion/deletion of 12-bp sequence (
Figure 3). Except for one of the SNPs (c.103C/T; p.Arg35Cys), all the other SNPs were either outside the coding region or did not lead to amino acid changes. The three-nucleotide sequence variation was located at c.83_c.85 and would result in two amino acid changes (p.Ser-Cys28_29Cys-Gly). The 12-bp insertion occurred in a tandem repeat of TGTGGCTA(T/C)GGC and led to variation in the number of repeats. There were three repeats in variant
E and two repeats in all the other variants. This additional repeat unit in variant
E would lead to the presence of an additional CGYG repeat in the central region of the protein (
Figure 4). Variant
B was notably different to all the other variants and possessed some unique nucleotide sequences that are observed in the ovine orthologue. In the coding region, variant
B was identical to ovine variants
B,
C, and
D (
Figure 3).
Phylogenetic analysis of the translated amino acid sequences revealed that the caprine
KRTAP6-2 sequences identified here were closer to ovine
KRTAP6-2 and ovine
KRTAP6-5, but different to other known HGT-
KRTAP sequences (
Figure 5). In contrast, the previously reported “caprine
KRTAP6-2” sequence (EU145019) was separate to all the clustered
KRTAP6-n sequences, but instead clustered with
KRTAP21-n, supporting the contention that the sequence of EU145010 is not caprine
KRTAP6-2.
The caprine KRTAP6-2 sequences would encode polypeptide with 83 and 87 amino acids, respectively. The polypeptides would contain a high content of glycine (38.6–40.2 mol %) and tyrosine (21.7–21.8 mol %) and a moderate level of cysteine (12.0–12.6 mol %) and serine (9.6–10.8 mol %). The caprine KAP6-2 proteins are predicted to be basic proteins with isoelectric point (pI) of 8.27–8.51.
3.3. Association of KRTAP6-2 Variation with Cashmere Fiber Traits
In the 230 goats investigated, 11 genotypes were detected, and these were: AA (n = 54), AB (n = 67), AC (n = 15), AD (n = 2.7), AE (n = 5), BB (n = 15), BC (n = 11), BD (n = 21), CC (n = 5), CD (n = 6), DD (n = 3), and EE (n = 1). This gives variant frequencies of 48.3%, 28.0%, 9.1%, 13.0%, and 1.5% for A to E, respectively.
As variant
E was found in only six goats (with a frequency of less than 5%), these six goats were removed from the association study and the effect of variant
E on cashmere fiber traits was not analyzed given the potential for these goats to bias the analyses. In the presence/absence models, the presence of variant
D was found to be associated with decreased MFD (present: 13.26 ± 0.07 µm; absent: 13.55 ± 0.04 µm;
p < 0.001) (
Table 1). No association was revealed with crimped fiber length or cashmere yield.
4. Discussion
This study describes the identification of a new caprine
KRTAP, sequence variation in the gene and an association between this variation and cashmere MFD in Longdong cashmere goats. The chromosomal location of this newly identified
KRTAP sequence matches well with the location of
KRTAP6-2 reported in sheep [
6], and the flanking sequences of this
KRTAP were more closely related to ovine
KRTAP6-2 flanking regions than any other ovine
KRTAP6-n, suggesting that the newly identified
KRTAP represent caprine
KRTAP6-2. The caprine
KRTAP6-2 is predicted to encode a basic protein that contains over 60 mol % of glycine and tyrosine, and this is consistent with the characteristics of an HGT-KAP and the observation that in sheep all known HGT-KAPs are basic proteins, except for KAP8-2 [
16].
While the number of variants and the nature of SNPs detected for caprine
KRTAP6-2 is comparable to that reported for its sheep orthologue [
17], some unique features are observed for caprine
KRTAP6-2. First is the high density of SNPs. The identification of 11 SNPs in the amplified region of 389-bp (excluding the primer binding regions) gives a density of 28.2 SNPs per kb. While
KRTAPs tend to have high density of SNPs [
6], the SNP density observed is much higher than its ovine orthologue, and higher than all the
KRTAPs identified in sheep except
KRTAP1-3.
The presence of a three-nucleotide sequence variation in the coding sequence of caprine
KRTAP6-2 is also notable. Despite this kind of variation (called trinucleotide polymorphisms; TNPs) being frequently found in the human genome, they are almost completely absent from coding exons and there are only three coding TNPs described in the Chinese human genome sequence and six from the Venter genome sequence [
25]. In humans, coding TNPs are reported for
KRTAP10-1 [
25], but these TNPs only lead to one amino acid change, whereas the TNP described here in caprine
KRTAP6-2 would lead to a change of two amino acids. The functional effect of the goat
KRTAP6-2 TNP awaits further investigation, but the lack of association of variant
B with cashmere traits (
Table 1) suggests that this TNP is unlikely to have an effect on fiber diameter and cashmere weight.
There is also length variation in the novel
KRTAP6-2 sequences. Given that all the variants were the same length except for variant
E which had an extra 12-bp in the tandem repeat region, it is likely that the length variation detected is due to the insertion of the 12-bp repeat in variant
E, rather than the deletion of the 12-bp repeat in all the other caprine
KRTAP6-2 variants. In sheep, length variation has been described for
KRTAP6-1,
KRTAP6-3, and
KRTAP6-5, but it has not been detected for
KRTAP6-2 and
KRTAP6-4 [
17,
26].
The variant
B sequence appears to have diverged from the other caprine variants to be more like the ovine
KRTAP6-2 variants, most notably being identical to some of the ovine variants in the coding region (
Figure 3). This phenomenon has not been reported for any other
KRTAPs, and its genesis is therefore uncertain. Given that the sequence variation reported here was found in only 230 Longdong goats from a single farm, more variants and additional variation may be found when more goats from different farms are investigated. This is supported by the differences observed between the sequences reported here and the goat genome assembly sequence NC_030808.1.
Caprine
KRTAP6-2 is clustered with
KRTAP20-1 and
KRTAP20-2 and located between them on chromosome 1, but the association detected
KRTAP6-2 variation and MFD is different to that reported for the nearby genes. Previous research with Longdong goats has revealed that variation in
KRTAP20-1 and
KRTAP20-2 are associated with cashmere fiber weight and crimped fiber length, but not MFD [
1,
12]. This suggests that the MFD association detected for
KRTAP6-2 may be because of
KRTAP6-2 itself and not because of
KRTAP20-1 and
KRTAP20-2, but in sheep, there are additional
KRTAPs clustered with
KRTAP6-2 in the region between
KRTAP20-1 and
KRTAP20-2—including
KRTAP6-1,
KRTAP6-3,
KRTAP6-4,
KRTAP6-5, and
KRTAP22-1 [
6]. Further investigation of these genes in goats and whether they associate with cashmere fiber traits may provide more information on the role of
KRTAP6-2 in fiber traits and the other genes too.
The detection of association between variant
D and cashmere MFD suggests variation in caprine affects the trait. Given that the association was only detected for variant
D, but not
A, and that these two variants differed by a single SNP (c.-20C/T) in the 5′-UTR and two synonymous SNPs (c.159C/T and c.189T/C) in the coding region, the effect detected for variant
D may be due to some of or all these SNPs. Despite not causing amino acid sequence changes, SNPs within the 5′-UTR, or synonymous SNPs in coding regions may nevertheless affect mRNA translation efficiency [
27].
The MFD of cashmere is an economically important trait. The associations found in this study suggest that caprine KRTAP6-2 may have potential as a gene-marker for selection for cashmere fiber traits that are of greater value in cashmere production.