Chicken Protein S Gene Regulates Adipogenesis and Affects Abdominal Fat Deposition
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experiments and Ethics Statement
2.2. Cell Culture
2.3. Reverse Transcription Reaction and Quantified Real-Time PCR (qRT-PCR)
2.4. Plasmid Construction and siRNA Synthesis
2.5. Western Blot
2.6. Polymorphisms in PROS1 and Their Associations with Growth Traits
2.7. Data Statistics and Analysis
3. Results
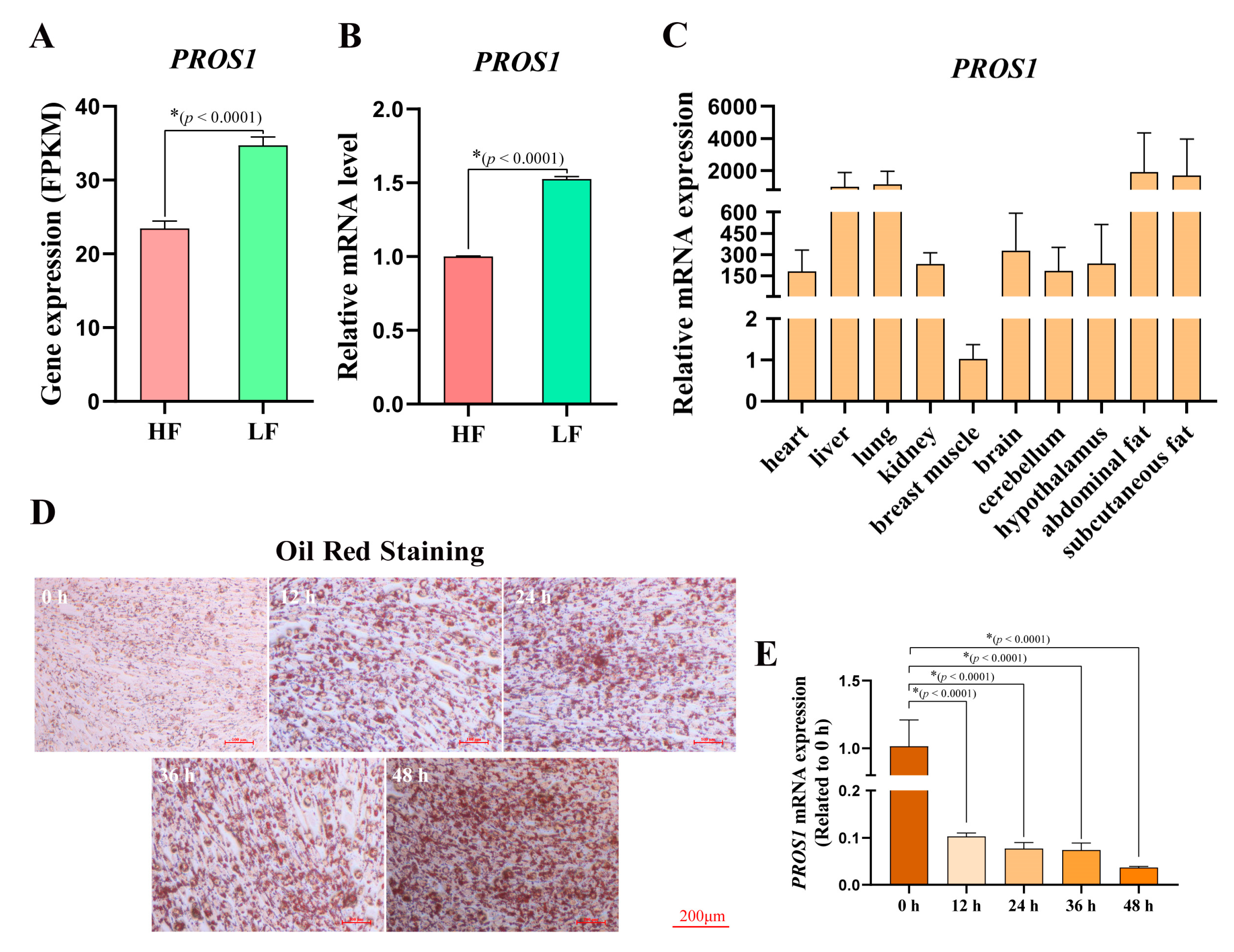
3.1. Aberrant Expression of PROS1 in Broilers with High Abdominal Fat Rate
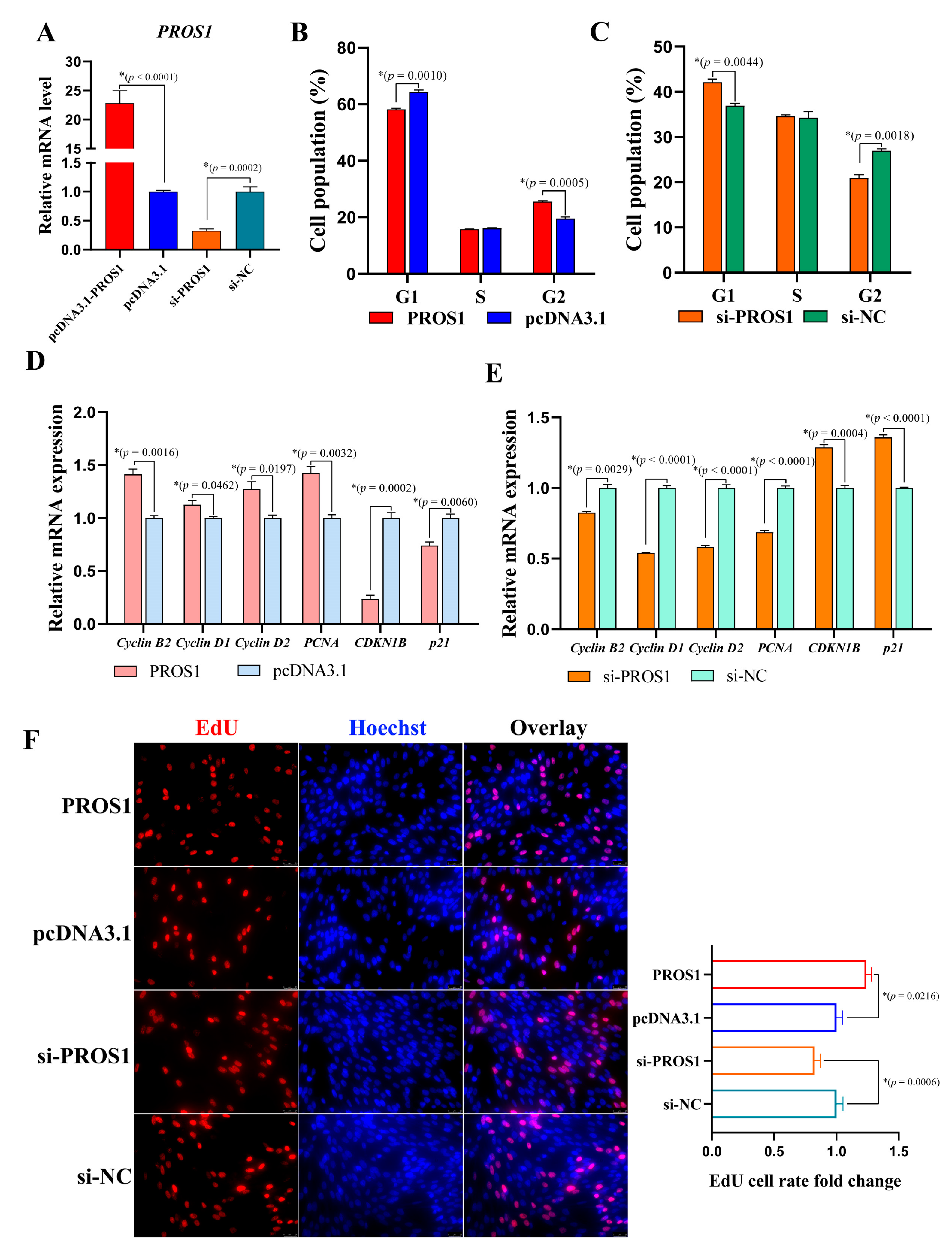
3.2. PROS1 Contributes to Chicken Preadipocyte Proliferation
3.3. PROS1 Suppresses Chicken Preadipocyte Differentiation
3.4. Association between SNPs in PROS1 5′ Flanking Region and Chicken Growth Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burt, D.W. The chicken genome. Genome Dyn. 2006, 2, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Furlong, R.F. Insights into vertebrate evolution from the chicken genome sequence. Genome Biol. 2005, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Consortium, I.C.G.S. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef]
- Wang, G.; Kim, W.K.; Cline, M.A.; Gilbert, E.R. Factors affecting adipose tissue development in chickens: A review. Poult. Sci. 2017, 96, 3687–3699. [Google Scholar] [CrossRef] [PubMed]
- Nematbakhsh, S.; Pei, C.P.; Selamat, J.; Nordin, N.; Idris, L.; Razis, A.A. Molecular regulation of lipogenesis, adipogenesis and fat deposition in chicken. Genes 2021, 12, 414. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S.; Kobayashi, M.; Murai, A.; Tsudzuki, M.; Ishikawa, A. Characterization of growth, fat deposition, and lipid Metabolism-Related gene expression in lean and obese Meat-Type chickens. J. Poult. Sci. 2019, 56, 101–111. [Google Scholar] [CrossRef]
- Fouad, A.M.; El-Senousey, H.K. Nutritional factors affecting abdominal fat deposition in poultry: A review. Asian-Australas J. Anim. Sci. 2014, 27, 1057–1068. [Google Scholar] [CrossRef]
- Zhang, M.; Li, F.; Ma, X.-F.; Li, W.-T.; Jiang, R.-R.; Han, R.-L.; Li, G.-X.; Wang, Y.-B.; Li, Z.-Y.; Tian, Y.-D.; et al. Identification of differentially expressed genes and pathways between intramuscular and abdominal fat-derived preadipocyte differentiation of chickens in vitro. BMC Genom. 2019, 20, 743. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chao, X.; Huang, W.; Li, Z.; Luan, K.; Ye, M.; Zhang, S.; Liu, M.; Li, H.; Luo, W.; et al. Whole transcriptome analysis reveals a potential regulatory mechanism of LncRNA-FNIP2/miR-24-3p/FNIP2 axis in chicken adipogenesis. Front. Cell Dev. Biol. 2021, 9, 653798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cai, B.; Ma, M.; Luo, W.; Zhang, Z.; Zhang, X.; Nie, Q. ALDH1A1 inhibits chicken preadipocytes’ proliferation and differentiation via the PPARgamma pathway in vitro and in vivo. Int. J. Mol. Sci. 2020, 21, 3150. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, S.; Ding, R.; Li, H.; Yang, C.-X.; Du, Z.-Q. Identification of a long noncoding RNA (lncPRDM16) inhibiting preadipocyte proliferation in the chicken. J. Agric. Food Chem. 2022, 70, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Borgel, D.; Gandrille, S.; Aiach, M. Protein S deficiency. Thromb. Haemost. 1997, 78, 351–356. [Google Scholar] [CrossRef]
- Schwarz, H.P.; Muntean, W.; Watzke, H.; Richter, B.; Griffin, J.H. Low total protein S antigen but high protein S activity due to decreased C4b-binding protein in neonates. Blood 1988, 71, 562–565. [Google Scholar] [CrossRef]
- Rubio-Aliaga, I.; Marvin-Guy, L.F.; Wang, P.; Wagnière, S.; Mansourian, R.; Fuerholz, A.; Saris, W.H.M.; Astrup, A.; Mariman, E.C.M.; Kussmann, M. Mechanisms of weight maintenance under high- and low-protein, low-glycaemic index diets. Mol. Nutr. Food Res. 2011, 55, 1603–1612. [Google Scholar] [CrossRef]
- Chao, X.; Guo, L.; Wang, Q.; Huang, W.; Liu, M.; Luan, K.; Jiang, J.; Lin, S.; Nie, Q.; Luo, W.; et al. MiR-429-3p/LPIN1 Axis Promotes Chicken Abdominal Fat Deposition via PPARgamma Pathway. Front. Cell Dev. Biol. 2020, 8, 595637. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, B.A.; Chen, J.; Nie, Q.; Zhang, X. Genomic insights into the multiple factors controlling abdominal fat deposition in a chicken model. Front. Genet. 2018, 9, 262. [Google Scholar] [CrossRef]
- Saller, F.; Brisset, A.C.; Tchaikovski, S.N.; Azevedo, M.; Chrast, R.; Fernández, J.A.; Schapira, M.; Hackeng, T.M.; Griffin, J.H.; Angelillo-Scherrer, A. Generation and phenotypic analysis of protein S-deficient mice. Blood 2009, 114, 2307–2314. [Google Scholar] [CrossRef] [PubMed]
- Burstyn-Cohen, T.; Heeb, M.J.; Lemke, G. Lack of protein S in mice causes embryonic lethal coagulopathy and vascular dysgenesis. J. Clin. Investig. 2009, 119, 2942–2953. [Google Scholar] [CrossRef]
- Resnyk, C.W.; Carré, W.; Wang, X.; Porter, T.E.; Simon, J.; le Bihan-Duval, E.; Duclos, M.J.; Aggrey, S.E.; Cogburn, L.A. Transcriptional analysis of abdominal fat in genetically fat and lean chickens reveals adipokines, lipogenic genes and a link between hemostasis and leanness. BMC Genom. 2013, 14, 557. [Google Scholar] [CrossRef]
- De Lange, M.; Snieder, H.; Ariëns, R.A.; Spector, T.D.; Grant, P.J. The genetics of haemostasis: A twin study. Lancet 2001, 357, 101–105. [Google Scholar] [CrossRef]
- Han, C.; Wang, J.; Li, L.; Wang, L.; Zhang, Z. The role of LXR alpha in goose primary hepatocyte lipogenesis. Mol. Cell. Biochem. 2009, 322, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Leveille, G.A.; Romsos, D.R.; Yeh, Y.-Y.; O’Hea, E.K. Lipid biosynthesis in the chick. A consideration of site of synthesis, influence of diet and possible regulatory mechanisms. Poult. Sci. 1975, 54, 1075–1093. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ren, X.; Li, L.; Lu, S.; Chen, T.; Tan, L.; Liu, M.; Luo, Q.; Liang, S.; Nie, Q.; et al. Integrative analyses of mRNA expression profile reveal the involvement of IGF2BP1 in chicken adipogenesis. Int. J. Mol. Sci. 2019, 20, 2923. [Google Scholar] [CrossRef]
- Resnyk, C.W.; Chen, C.; Huang, H.; Wu, C.H.; Simon, J.; Le Bihan-Duval, E.; Duclos, M.J.; Cogburn, L. A RNA-Seq analysis of abdominal fat in genetically fat and lean chickens highlights a divergence in expression of genes controlling adiposity, hemostasis, and lipid metabolism. PLoS ONE 2015, 10, e139549. [Google Scholar] [CrossRef] [PubMed]
- Maimon, A.; Levi-Yahid, V.; Ben-Meir, K.; Halpern, A.; Talmi, Z.; Priya, S.; Mizraji, G.; Mistriel-Zerbib, S.; Berger, M.; Baniyash, M.; et al. Myeloid cell-derived PROS1 inhibits tumor metastasis by regulating inflammatory and immune responses via IL-10. J. Clin. Investig. 2021, 131, e126089. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Liu, R.; Zhao, G.; Zheng, M.; Chen, J.; Wen, J. Folate supplementation modifies CCAAT/enhancer-binding protein alpha methylation to mediate differentiation of preadipocytes in chickens. Poult. Sci. 2014, 93, 2596–2603. [Google Scholar] [CrossRef]
- Wang, L.; NA, W.; Wang, Y.-X.; Wang, Y.-B.; Wang, N.; Wang, Q.-G.; Li, Y.-M.; Li, H. Characterization of chicken PPARgamma expression and its impact on adipocyte proliferation and differentiation. Yi Chuan 2012, 34, 454–464. [Google Scholar] [CrossRef]
- Sun, Y.N.; Gao, Y.; Qiao, S.P.; Wang, S.Z.; Duan, K.; Wang, Y.X.; Li, H.; Wang, N. Epigenetic DNA methylation in the promoters of peroxisome proliferator-activated receptor gamma in chicken lines divergently selected for fatness. J. Anim. Sci. 2014, 92, 48–53. [Google Scholar] [CrossRef]
- Chen, S.; Li, K.; Cao, W.; Wang, J.; Zhao, T.; Huan, Q.; Yang, Y.-F.; Wu, S.; Qian, W. Codon-Resolution analysis reveals a direct and Context-Dependent impact of individual synonymous mutations on mRNA level. Mol. Biol. Evol. 2017, 34, 2944–2958. [Google Scholar] [CrossRef]
- Hunt, R.; Sauna, Z.E.; Ambudkar, S.V.; Gottesman, M.M.; Kimchi-Sarfaty, C. Silent (synonymous) SNPs: Should we care about them? Methods Mol. Biol. 2009, 578, 23–39. [Google Scholar] [CrossRef]

| SNPs | Position in Genome | Allele | Genotypes of Alleles | Allelic Frequency | p-Value | ||
|---|---|---|---|---|---|---|---|
| rs740370863 | NC_006088.5: 91131134 | T→C | TT (n = 146) | TC (n = 127) | CC (n = 15) | T = 0.727 C = 0.273 | 0.057 |
| rs731399734 | NC_006088.5: 91131136 | T→C | TT (n = 50) | TC (n = 144) | CC (n = 194) | T = 0.424 C = 0.576 | 0.685 |
| rs736154944 | NC_006088.5: 91131140 | G→T | GG (n = 145) | GT (n = 128) | TT (n = 15) | G = 0.726 T = 0.274 | 0.048 |
| rs739467563 | NC_006088.5: 91131187 | A→T | AA (n = 148) | AT (n = 127) | TT (n = 13) | A = 0.734 T = 0.266 | 0.027 |
| rs317681540 | NC_006088.5: 91131298 | T→C | TT (n = 13) | TC (n = 111) | CC (n = 164) | T = 0.238 C = 0.762 | 0.284 |
| rs315810115 | NC_006088.5: 91131319 | G→A | GG (n = 13) | GA (n = 111) | AA (n = 164) | T = 0.238 C = 0.762 | 0.284 |
| rs313239277 | NC_006088.5: 91131349 | G→A | GG (n = 10) | GA (n = 106) | AA (n = 172) | G = 0.219 A = 0.781 | 0.192 |
| rs317295547 | NC_006088.5: 91131410 | G→A | GG (n = 144) | GA (n = 126) | AA (n = 18) | G = 0.719 A = 0.281 | 0.163 |
| Traits | Genotypes (Number) | p-Value | ||
|---|---|---|---|---|
| rs740370863 | CC (n = 15) | TC (n = 127) | TT (n = 146) | |
| IFW | 24.33 ± 10.66 ab | 24.40 ± 10.10 a | 28.13 ± 9.73 b | 0.007 |
| AFW | 24.83 ± 12.59 a | 29.79 ± 16.73 a | 39.31 ± 15.93 b | 0.000 |
| AFWR | 1.87 ± 0.91 a | 2.30 ± 1.22 a | 3.07 ± 1.215 b | 0.000 |
| rs731399734 | CC (n = 94) | TC (n = 144) | TT (n = 50) | |
| AFW | 37.31 ± 15.30 a | 33.67 ± 18.14 ab | 30.80 ± 15.33 b | 0.069 |
| AFWR | 2.93 ± 1.11 a | 2.61 ± 1.39 ab | 2.34 ± 1.09 b | 0.022 |
| rs736154944 | GG (n = 145) | GT (n = 128) | TT (n = 15) | |
| IFW | 28.14 ± 9.66 a | 24.41 ± 10.18 b | 24.33 ± 10.66 ab | 0.007 |
| AFW | 39.48 ± 15.95 a | 29.67 ± 16.61 b | 24.83 ± 12.59 b | 0.000 |
| AFWR | 3.08 ± 1.22 a | 2.29 ± 1.21 b | 1.87 ± 0.91 b | 0.000 |
| rs739467563 | AA (n = 148) | AT (n = 127) | TT (n = 13) | |
| FEW | 1246.18 ± 169.95 a | 1265.13 ± 151.95 ab | 1346.32 ± 191.57 b | 0.092 |
| HEW | 1493.74 ± 196.33 a | 1509.96 ± 177.35 ab | 1605.09 ± 227.01 b | 0.123 |
| SL | 75.06 ± 5.14 a | 76.36 ± 5.54 b | 76.92 ± 4.79 ab | 0.093 |
| IFW | 27.81 ± 9.74 a | 24.53 ± 10.24 b | 26.02 ± 10.37 ab | 0.026 |
| AFW | 38.81 ± 15.74 a | 30.02 ± 17.18 b | 26.03 ± 12.98 b | 0.000 |
| AFWR | 3.04 ± 1.21 a | 2.31 ± 1.23 b | 1.94 ± 0.97 b | 0.000 |
| rs317681540 | CC (n = 164) | TC (n = 111) | TT (n = 13) | |
| FEW | 1246.18 ± 169.95 a | 1265.13 ± 151.95 ab | 1346.32 ± 191.57 b | 0.092 |
| HEW | 1493.74 ± 196.33 a | 1509.96 ± 177.35 ab | 1605.09 ± 227.01 b | 0.123 |
| SL | 75.06 ± 5.14 a | 76.36 ± 5.54 b | 76.92 ± 4.79 ab | 0.093 |
| IFW | 27.81 ± 9.74 a | 24.53 ± 10.24 b | 26.02 ± 10.37 ab | 0.026 |
| AFW | 38.81 ± 15.74 a | 30.02 ± 17.18 b | 26.03 ± 12.98 b | 0.000 |
| AFWR | 3.04 ± 1.21 a | 2.31 ± 1.23 b | 1.94 ± 0.97 b | 0.000 |
| rs313239277 | AA (n = 176) | GA (n = 106) | GG (n = 10) | |
| AFWR | 2.53 ± 1.22 a | 2.87 ± 1.32 b | 2.81 ± 1.31 b | 0.094 |
| rs317295547 | AA (n = 18) | GA (n = 126) | GG (n = 144) | |
| IFW | 25.80 ± 10.45 ab | 24.15 ± 10.03 a | 28.21 ± 9.77 b | 0.004 |
| AFW | 24.82 ± 11.84 a | 29.82 ± 16.76 a | 39.52 ± 15.93 b | 0.000 |
| AFWR | 1.90 ± 0.89 a | 2.30 ± 1.22 a | 3.08 ± 1.22 b | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, L.; Huang, W.; Zhang, S.; Huang, Y.; Xu, Y.; Wu, R.; Fang, X.; Xu, H.; Nie, Q. Chicken Protein S Gene Regulates Adipogenesis and Affects Abdominal Fat Deposition. Animals 2022, 12, 2046. https://doi.org/10.3390/ani12162046
Guo L, Huang W, Zhang S, Huang Y, Xu Y, Wu R, Fang X, Xu H, Nie Q. Chicken Protein S Gene Regulates Adipogenesis and Affects Abdominal Fat Deposition. Animals. 2022; 12(16):2046. https://doi.org/10.3390/ani12162046
Chicago/Turabian StyleGuo, Lijin, Weiling Huang, Siyu Zhang, Yulin Huang, Yibin Xu, Ruiquan Wu, Xiang Fang, Haiping Xu, and Qinghua Nie. 2022. "Chicken Protein S Gene Regulates Adipogenesis and Affects Abdominal Fat Deposition" Animals 12, no. 16: 2046. https://doi.org/10.3390/ani12162046






