The World of Organoids: Gastrointestinal Disease Modelling in the Age of 3R and One Health with Specific Relevance to Dogs and Cats
Abstract
:Simple Summary
Abstract
1. Introduction
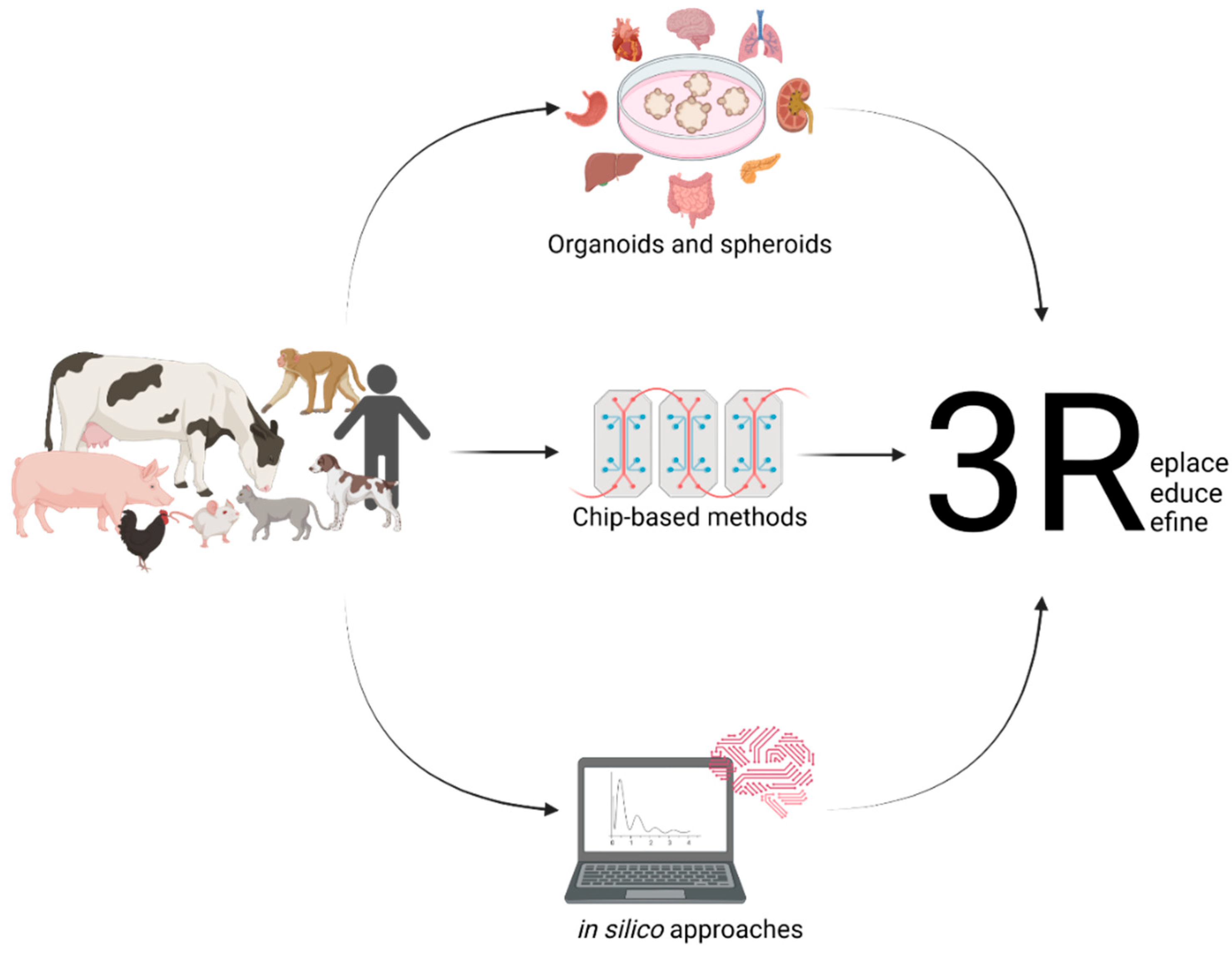
2. The Importance of Organoids for One Health
3. Organoids Modelling the Intestinal Epithelium
3.1. Microinjection
3.2. Apical-Out Organoids
3.3. Organoid-Derived Monolayers
4. Limitations
5. Outlook
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anholt, M.; Barkema, H. What Is One Health? Can Vet. J. 2021, 62, 641–644. [Google Scholar] [PubMed]
- Nerpel, A.; Yang, L.; Sorger, J.; Käsbohrer, A.; Walzer, C.; Desvars-Larrive, A. SARS-ANI: A Global Open Access Dataset of Reported SARS-CoV-2 Events in Animals. Sci Data 2022, 9, 438. [Google Scholar] [CrossRef] [PubMed]
- Amman, F.; Markt, R.; Endler, L.; Hupfauf, S.; Agerer, B.; Schedl, A.; Richter, L.; Zechmeister, M.; Bicher, M.; Heiler, G.; et al. Viral Variant-Resolved Wastewater Surveillance of SARS-CoV-2 at National Scale. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Russell, W.; Burch, R. The Principles of Humane Experimental Technique. In John Hopkins Bloomberg School of Public Health; 1959; Available online: https://caat.jhsph.edu/principles/the-principles-of-humane-experimental-technique (accessed on 9 September 2022).
- Office of the Law Revision Counsel. Title 7 of the Code of Laws of the United States of America, Chapter 54: Transportation, Sale, and Handling of Certain Animals; US Congress. Available online: http://uscode.house.gov/view.xhtml?path=/prelim@title7/chapter54&edition=prelim (accessed on 9 September 2022).
- European Parliament; European Council. Directive 2010/63/EU; European Union, 2010. Available online: https://norecopa.no/legislation/eu-directive-201063 (accessed on 9 September 2022).
- Max Planck Society for the Advancement of Science, e.V. Reduce, Refine, Replace–Responsibility. Available online: https://www.mpg.de/10973438/4rs (accessed on 4 July 2022).
- European Commission. Summary Report on the Statistics on the Use of Animals for Scientific Purposes in the Member States of the European Union and Norway in 2018; Brussels, 2021. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwj2nInjn536AhVCumMGHXqaCcEQFnoECA0QAQ&url=https%3A%2F%2Fec.europa.eu%2Fenvironment%2Fchemicals%2Flab_animals%2Fpdf%2FSWD_%2520part_A_and_B.pdf&usg=AOvVaw0opxqbviYJ0WzPyGdJUFKo (accessed on 9 September 2022).
- United States Department of Agriculture. Annual Report Animal Usage by Fiscal Year. Available online: https://Www.Aphis.Usda.Gov/Animal_welfare/Downloads/Reports/Fy19-Summary-Report-Column-F.Pdf (accessed on 4 July 2022).
- Ellahham, S. Artificial Intelligence: The Future for Diabetes Care. Am. J. Med. 2020, 133, 895–900. [Google Scholar] [CrossRef] [PubMed]
- Mohd Faizal, A.S.; Thevarajah, T.M.; Khor, S.M.; Chang, S.W. A Review of Risk Prediction Models in Cardiovascular Disease: Conventional Approach vs. Artificial Intelligent Approach. Comput Methods Programs Biomed 2021, 207, 106190. [Google Scholar] [CrossRef] [PubMed]
- Peetluk, L.S.; Ridolfi, F.M.; Rebeiro, P.F.; Liu, D.; Rolla, V.C.; Sterling, T.R. Systematic Review of Prediction Models for Pulmonary Tuberculosis Treatment Outcomes in Adults. BMJ Open 2021, 11, e044687. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Luna, J.; Grisoni, F.; Weskamp, N.; Schneider, G. Artificial Intelligence in Drug Discovery: Recent Advances and Future Perspectives. Expert Opin. Drug Discov. 2021, 16, 949–959. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Devi, G.R. Three-Dimensional Culture Systems in Cancer Research: Focus on Tumor Spheroid Model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Willyard, C. Rise of the Organoids. Nature 2015, 523, 1–10. [Google Scholar]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowan, C.S.; Renner, M.; de Gennaro, M.; Gross-Scherf, B.; Goldblum, D.; Hou, Y.; Munz, M.; Rodrigues, T.M.; Krol, J.; Szikra, T.; et al. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell 2020, 182, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.Y.; Choi, W.H.; Jeon, S.G.; Lee, S.; Park, J.M.; Park, M.; Lee, H.; Lew, H.; Yoo, J. Establishment of Functional Epithelial Organoids from Human Lacrimal Glands. Stem Cell Res. Ther. 2021, 12, 247. [Google Scholar] [CrossRef] [PubMed]
- Ogundipe, V.M.L.; Groen, A.H.; Hosper, N.; Nagle, P.W.K.; Hess, J.; Faber, H.; Jellema, A.L.; Baanstra, M.; Links, T.P.; Unger, K.; et al. Generation and Differentiation of Adult Tissue-Derived Human Thyroid Organoids. Stem Cell Rep. 2021, 16, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Wimmer, R.A.; Leopoldi, A.; Aichinger, M.; Wick, N.; Hantusch, B.; Novatchkova, M.; Taubenschmid, J.; Hämmerle, M.; Esk, C.; Bagley, J.A.; et al. Human Blood Vessel Organoids as a Model of Diabetic Vasculopathy. Nature 2019, 565, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Israeli, Y.R.; Wasserman, A.H.; Gabalski, M.A.; Volmert, B.D.; Ming, Y.; Ball, K.A.; Yang, W.; Zou, J.; Ni, G.; Pajares, N.; et al. Self-Assembling Human Heart Organoids for the Modeling of Cardiac Development and Congenital Heart Disease. Nat. Commun. 2021, 12, 5142. [Google Scholar] [CrossRef]
- Dekkers, J.F.; van Vliet, E.J.; Sachs, N.; Rosenbluth, J.M.; Kopper, O.; Rebel, H.G.; Wehrens, E.J.; Piani, C.; Visvader, J.E.; Verissimo, C.S.; et al. Long-Term Culture, Genetic Manipulation and Xenotransplantation of Human Normal and Breast Cancer Organoids. Nat. Protoc. 2021, 16, 1936–1965. [Google Scholar] [CrossRef]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Takasato, M.; Er, P.X.; Chiu, H.S.; Maier, B.; Baillie, G.J.; Ferguson, C.; Parton, R.G.; Wolvetang, E.J.; Roost, M.S.; de Sousa Lopes, S.M.C.; et al. Kidney Organoids from Human IPS Cells Contain Multiple Lineages and Model Human Nephrogenesis. Nature 2015, 526, 564–568. [Google Scholar] [CrossRef] [PubMed]
- Broutier, L.; Andersson-Rolf, A.; Hindley, C.J.; Boj, S.F.; Clevers, H.; Koo, B.K.; Huch, M. Culture and Establishment of Self-Renewing Human and Mouse Adult Liver and Pancreas 3D Organoids and Their Genetic Manipulation. Nat. Protoc. 2016, 11, 1724–1743. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; van Es, J.H.; van den Brink, S.; van Houdt, W.J.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-Term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 Stem Cells Build Crypt-Villus Structures in Vitro without a Mesenchymal Niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef]
- Chang, Y.H.; Chu, T.Y.; Ding, D.C. Human Fallopian Tube Epithelial Cells Exhibit Stemness Features, Self-Renewal Capacity, and Wnt-Related Organoid Formation. J Biomed Sci 2020, 27, 32. [Google Scholar] [CrossRef] [PubMed]
- Boretto, M.; Cox, B.; Noben, M.; Hendriks, N.; Fassbender, A.; Roose, H.; Amant, F.; Timmerman, D.; Tomassetti, C.; Vanhie, A.; et al. Development of Organoids from Mouse and Human Endometrium Showing Endometrial Epithelium Physiology and Long-Term Expandability. Development (Cambridge) 2017, 144, 1775–1786. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Choi, S.; Kang, B.; Kong, J.; Kim, Y.; Yoon, W.H.; Lee, H.-R.; Kim, S.; Kim, H.-M.; Lee, H.; et al. Creation of Bladder Assembloids Mimicking Tissue Regeneration and Cancer. Nature 2020, 588, 664–669. [Google Scholar] [CrossRef]
- Drost, J.; Karthaus, W.R.; Gao, D.; Driehuis, E.; Sawyers, C.L.; Chen, Y.; Clevers, H. Organoid Culture Systems for Prostate Epithelial and Cancer Tissue. Nat. Protoc. 2016, 11, 347–358. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988. [Google Scholar] [CrossRef] [PubMed]
- Chandra, L.; Borcherding, D.C.; Kingsbury, D.; Atherly, T.; Ambrosini, Y.M.; Bourgois-Mochel, A.; Yuan, W.; Kimber, M.; Qi, Y.; Wang, Q.; et al. Derivation of Adult Canine Intestinal Organoids for Translational Research in Gastroenterology. BMC Biol. 2019, 17, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kramer, N.; Pratscher, B.; Meneses, A.M.C.; Tschulenk, W.; Walter, I.; Swoboda, A.; Kruitwagen, H.S.; Schneeberger, K.; Penning, L.C.; Spee, B.; et al. Generation of Differentiating and Long-Living Intestinal Organoids Reflecting the Cellular Diversity of Canine Intestine. Cells 2020, 9, 822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gabriel, V.; Zdyrski, C.; Sahoo, D.K.; Dao, K.; Bourgois-Mochel, A.; Kopper, J.; Zeng, X.L.; Estes, M.K.; Mochel, J.P.; Allenspach, K. Standardization and Maintenance of 3D Canine Hepatic and Intestinal Organoid Cultures for Use in Biomedical Research. J. Vis. Exp. 2022, 2022. [Google Scholar] [CrossRef]
- Tekes, G.; Ehmann, R.; Boulant, S.; Stanifer, M.L. Development of Feline Ileum- and Colon-Derived Organoids and Their Potential Use to Support Feline Coronavirus Infection. Cells 2020, 9, 2085. [Google Scholar] [CrossRef]
- Kruitwagen, H.S.; Oosterhoff, L.A.; Vernooij, I.G.W.H.; Schrall, I.M.; van Wolferen, M.E.; Bannink, F.; Roesch, C.; van Uden, L.; Molenaar, M.R.; Helms, J.B.; et al. Long-Term Adult Feline Liver Organoid Cultures for Disease Modeling of Hepatic Steatosis. Stem Cell Rep. 2017, 8, 822–830. [Google Scholar] [CrossRef]
- Kruitwagen, H.S.; Oosterhoff, L.A.; van Wolferen, M.E.; Chen, C.; Nantasanti Assawarachan, S.; Schneeberger, K.; Kummeling, A.; van Straten, G.; Akkerdaas, I.C.; Vinke, C.R.; et al. Long-Term Survival of Transplanted Autologous Canine Liver Organoids in a COMMD1-Deficient Dog Model of Metabolic Liver Disease. Cells 2020, 9, 410. [Google Scholar] [CrossRef]
- Chen, T.C.; Neupane, M.; Chien, S.J.; Chuang, F.R.; Crawford, R.B.; Kaminski, N.E.; Chang, C.C. Characterization of Adult Canine Kidney Epithelial Stem Cells That Give Rise to Dome-Forming Tubular Cells. Stem Cells Dev. 2019, 28, 1424–1433. [Google Scholar] [CrossRef]
- Elbadawy, M.; Usui, T.; Mori, T.; Tsunedomi, R.; Hazama, S.; Nabeta, R.; Uchide, T.; Fukushima, R.; Yoshida, T.; Shibutani, M.; et al. Establishment of a Novel Experimental Model for Muscle-Invasive Bladder Cancer Using a Dog Bladder Cancer Organoid Culture. Cancer Sci. 2019, 110, 2806–2821. [Google Scholar] [CrossRef]
- Usui, T.; Sakurai, M.; Nishikawa, S.; Umata, K.; Nemoto, Y.; Haraguchi, T.; Itamoto, K.; Mizuno, T.; Noguchi, S.; Mori, T.; et al. Establishment of a Dog Primary Prostate Cancer Organoid Using the Urine Cancer Stem Cells. Cancer Sci. 2017, 108, 2383–2392. [Google Scholar] [CrossRef]
- Wiener, D.J.; Basak, O.; Asra, P.; Boonekamp, K.E.; Kretzschmar, K.; Papaspyropoulos, A.; Clevers, H. Establishment and Characterization of a Canine Keratinocyte Organoid Culture System. Vet. Dermatol. 2018, 29, 375-e126. [Google Scholar] [CrossRef]
- Jankovic, J.; Dettwiler, M.; Fernández, M.G.; Tièche, E.; Hahn, K.; April-Monn, S.; Dettmer, M.S.; Kessler, M.; Rottenberg, S.; Campos, M. Validation of Immunohistochemistry for Canine Proteins Involved in Thyroid Iodine Uptake and Their Expression in Canine Follicular Cell Thyroid Carcinomas (FTCs) and FTC-Derived Organoids. Vet. Pathol. 2021, 58, 1172–1180. [Google Scholar] [CrossRef]
- Powell, R.H.; Behnke, M.S. WRN Conditioned Media Is Sufficient for in Vitro Propagation of Intestinal Organoids from Large Farm and Small Companion Animals. Biol. Open 2017, 6, 698–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawasaki, M.; Goyama, T.; Tachibana, Y.; Nagao, I.; Ambrosini, Y.M. Farm and Companion Animal Organoid Models in Translational Research: A Powerful Tool to Bridge the Gap Between Mice and Humans. Front. Med. Technol. 2022, 4, 895379. [Google Scholar] [CrossRef]
- Hoffmann, A.R.; Proctor, L.M.; Surette, M.G.; Suchodolski, J.S. The Microbiome: The Trillions of Microorganisms That Maintain Health and Cause Disease in Humans and Companion Animals. Vet. Pathol. 2016, 53, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Honneffer, J.B.; Minamoto, Y.; Suchodolski, J.S. Microbiota Alterations in Acute and Chronic Gastrointestinal Inflammation of Cats and Dogs. World J. Gastroenterol. 2014, 20, 16489–16497. [Google Scholar] [CrossRef]
- Mondo, E.; Marliani, G.; Accorsi, P.A.; Cocchi, M.; di Leone, A. Role of Gut Microbiota in Dog and Cat’s Health and Diseases. Open Vet. J. 2019, 9, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Sack, A.; Palanisamy, G.; Manuel, M.; Paulsamy, C.; Rose, A.; Kaliappan, S.P.; Ward, H.; Walson, J.L.; Halliday, K.E.; Rao Ajjampur, S.S. A One Health Approach to Defining Animal and Human Helminth Exposure Risks in a Tribal Village in Southern India. Am. J. Trop. Med. Hyg. 2021, 105, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Galán-Puchades, M.T.; Trelis, M.; Sáez-Durán, S.; Cifre, S.; Gosálvez, C.; Sanxis-Furió, J.; Pascual, J.; Bueno-Marí, R.; Franco, S.; Peracho, V.; et al. One Health Approach to Zoonotic Parasites: Molecular Detection of Intestinal Protozoans in an Urban Population of Norway Rats, Rattus Norvegicus, in Barcelona, Spain. Pathogens 2021, 10, 311. [Google Scholar] [CrossRef]
- Usui, M. Prevalence of Clostridioides Difficile in Animals and Its Relationship with Human Infections. Jpn. J. Chemother. 2020, 68, 557–562. [Google Scholar]
- Usui, M. One Health Approach to Clostridioides Difficile in Japan. J. Infect. Chemother. 2020, 26, 643–650. [Google Scholar] [CrossRef]
- Finsterwalder, S.K.; Loncaric, I.; Cabal, A.; Szostak, M.P.; Barf, L.M.; Marz, M.; Allerberger, F.; Burgener, I.A.; Tichy, A.; Feßler, A.T.; et al. Dogs as Carriers of Virulent and Resistant Genotypes of Clostridioides Difficile. Zoonoses Public Health 2022, 69, 673–681. [Google Scholar] [CrossRef]
- Ehling-Schulz, M.; Lereclus, D.; Koehler, T.M. The Bacillus Cereus Group: Bacillus Species with Pathogenic Potential. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.P.; Humphrey, T.J.; Maskell, D.J. Molecular Insights into Farm Animal and Zoonotic Salmonella Infections. Philos. Trans. R. Soc. B Biol. Sci. 2009, 364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez Diaz, C.; Seyboldt, C.; Rupnik, M. Non-Human C. Difficile Reservoirs and Sources: Animals, Food, Environment. Adv. Exp. Med. Biol. 2018, 1050, 227–243. [Google Scholar] [PubMed]
- McLure, A.; Clements, A.C.A.; Kirk, M.; Glass, K. Modelling Diverse Sources of Clostridium Difficile in the Community: Importance of Animals, Infants and Asymptomatic Carriers. Epidemiol. Infect. 2019, 147, E152. [Google Scholar] [CrossRef] [PubMed]
- Otten, A.M.; Reid-Smith, R.J.; Fazil, A.; Weese, J.S. Disease Transmission Model for Community-Associated Clostridium Difficile Infection. Epidemiol. Infect. 2010, 138, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Redding, L.E.; Tu, V.; Abbas, A.; Alvarez, M.; Zackular, J.P.; Gu, C.; Bushman, F.D.; Kelly, D.J.; Barnhart, D.; Lee, J.J.; et al. Genetic and Phenotypic Characteristics of Clostridium (Clostridioides) Difficile from Canine, Bovine, and Pediatric Populations. Anaerobe 2022, 74, 102539. [Google Scholar] [CrossRef]
- Bell, V.; Ferrão, J.; Pimentel, L.; Pintado, M.; Fernandes, T. One Health, Fermented Foods, and Gut Microbiota. Foods 2018, 7, 195. [Google Scholar] [CrossRef]
- Schulte, L.; Hohwieler, M.; Müller, M.; Klaus, J. Intestinal Organoids as a Novel Complementary Model to Dissect Inflammatory Bowel Disease. Stem Cells Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Kopper, J.J.; Iennarella-Servantez, C.; Jergens, A.E.; Sahoo, D.K.; Guillot, E.; Bourgois-Mochel, A.; Martinez, M.N.; Allenspach, K.; Mochel, J.P. Harnessing the Biology of Canine Intestinal Organoids to Heighten Understanding of Inflammatory Bowel Disease Pathogenesis and Accelerate Drug Discovery: A One Health Approach. Front. Toxicol. 2021, 3. [Google Scholar] [CrossRef]
- Gunasekera, S.; Zahedi, A.; O’dea, M.; King, B.; Monis, P.; Thierry, B.; Carr, J.M.; Ryan, U. Organoids and Bioengineered Intestinal Models: Potential Solutions to the Cryptosporidium Culturing Dilemma. Microorganisms 2020, 8, 715. [Google Scholar] [CrossRef]
- Bowcutt, R.; Forman, R.; Glymenaki, M.; Carding, S.R.; Else, K.J.; Cruickshank, S.M. Heterogeneity across the Murine Small and Large Intestine. World J. Gastroenterol. 2014, 20, 15216. [Google Scholar] [CrossRef] [PubMed]
- Cramer, J.M.; Thompson, T.; Geskin, A.; Laframboise, W.; Lagasse, E. Distinct Human Stem Cell Populations in Small and Large Intestine. PLoS ONE 2015, 10, e0118792. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Takahashi, D.; Takano, S.; Kimura, S.; Hase, K. The Roles of Peyer’s Patches and Microfold Cells in the Gut Immune System: Relevance to Autoimmune Diseases. Front. Immunol. 2019, 10, 2345. [Google Scholar] [CrossRef] [PubMed]
- Burclaff, J.; Bliton, R.J.; Breau, K.A.; Ok, M.T.; Gomez-Martinez, I.; Ranek, J.S.; Bhatt, A.P.; Purvis, J.E.; Woosley, J.T.; Magness, S.T. A Proximal-to-Distal Survey of Healthy Adult Human Small Intestine and Colon Epithelium by Single-Cell Transcriptomics. CMGH 2022, 13, 1554–1589. [Google Scholar] [CrossRef]
- Ghoos, Y.; Vantrappen, G. The Cytochemical Localization of Lysozyme in Paneth Cell Granules. Histochem. J. 1971, 3, 175–178. [Google Scholar] [CrossRef]
- Sheahan, D.G.; Jervis, H.R. Comparative Histochemistry of Gastrointestinal Mucosubstances. Am. J. Anat. 1976, 146, 103–131. [Google Scholar] [CrossRef]
- Co, J.Y.; Margalef-Català, M.; Li, X.; Mah, A.T.; Kuo, C.J.; Monack, D.M.; Amieva, M.R. Controlling Epithelial Polarity: A Human Enteroid Model for Host-Pathogen Interactions. Cell Rep. 2019, 26, 2509–2520.e4. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Zhang, M.; Wang, H.; Cui, A.; Zhao, J.; Ji, W.; Chen, Y.G. Establishment of Porcine and Monkey Colonic Organoids for Drug Toxicity Study. Cell Regen. 2021, 10, 32. [Google Scholar] [CrossRef]
- Rosselot, A.E.; Park, M.; Kim, M.; Matsu-Ura, T.; Wu, G.; Flores, D.E.; Subramanian, K.R.; Lee, S.; Sundaram, N.; Broda, T.R.; et al. Ontogeny and Function of the Circadian Clock in Intestinal Organoids. EMBO.J 2022, 41, e106973. [Google Scholar] [CrossRef]
- Hill, D.R.; Huang, S.; Nagy, M.S.; Yadagiri, V.K.; Fields, C.; Mukherjee, D.; Bons, B.; Dedhia, P.H.; Chin, A.M.; Tsai, Y.H.; et al. Bacterial Colonization Stimulates a Complex Physiological Response in the Immature Human Intestinal Epithelium. Elife 2017, 6, e29132. [Google Scholar] [CrossRef]
- Abuaita, B.H.; Lawrence, A.L.E.; Berger, R.P.; Hill, D.R.; Huang, S.; Yadagiri, V.K.; Bons, B.; Fields, C.; Wobus, C.E.; Spence, J.R.; et al. Comparative Transcriptional Profiling of the Early Host Response to Infection by Typhoidal and Non-Typhoidal Salmonella Serovars in Human Intestinal Organoids. PLoS Pathog. 2021, 17, e1009987. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Heo, I.; O’connor, R. Studying Cryptosporidium Infection in 3D Tissue-Derived Human Organoid Culture Systems by Microinjection. J. Vis. Exp. 2019, 2019, e59610. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.S.; Ki, S.J.; Thanavel, R.; Kim, J.J.; Lee, M.O.; Kim, J.; Jung, C.R.; Han, T.S.; Cho, H.S.; Ryu, C.M.; et al. Maturation of Human Intestinal Organoids in Vitro Facilitates Colonization by Commensal Lactobacilli by Reinforcing the Mucus Layer. FASEB J. 2020, 34, 9899–9910. [Google Scholar] [CrossRef]
- Stroulios, G.; Stahl, M.; Elstone, F.; Chang, W.; Louis, S.; Eaves, A.; Simmini, S.; Conder, R.K. Culture Methods to Study Apical-Specific Interactions Using Intestinal Organoid Models. J. Vis. Exp. 2021, 2021, e62330. [Google Scholar] [CrossRef] [PubMed]
- Ojakian, G.K.; Schwimmer, R. Regulation of Epithelial Cell Surface Polarity Reversal by Β1 Integrins. J. Cell Sci. 1994, 107 Pt 3, 561–576.e6. [Google Scholar] [CrossRef]
- Joo, S.S.; Gu, B.H.; Park, Y.J.; Rim, C.Y.; Kim, M.J.; Kim, S.H.; Cho, J.H.; Kim, H.B.; Kim, M. Porcine Intestinal Apical-Out Organoid Model for Gut Function Study. Animals 2022, 12, 372. [Google Scholar] [CrossRef]
- Li, Y.; Yang, N.; Chen, J.; Huang, X.; Zhang, N.; Yang, S.; Liu, G.; Liu, G. Next-Generation Porcine Intestinal Organoids: An Apical-Out Organoid Model for Swine Enteric Virus Infection and Immune Response Investigations. J. Virol. 2020, 94, e01006–e01020. [Google Scholar] [CrossRef]
- Smith, D.; Price, D.R.G.; Burrells, A.; Faber, M.N.; Hildersley, K.A.; Chintoan-Uta, C.; Chapuis, A.F.; Stevens, M.; Stevenson, K.; Burgess, S.T.G.; et al. The Development of Ovine Gastric and Intestinal Organoids for Studying Ruminant Host-Pathogen Interactions. Front Cell Infect Microbiol. 2021, 11. [Google Scholar] [CrossRef]
- Nash, T.J.; Morris, K.M.; Mabbott, N.A.; Vervelde, L. Inside-out Chicken Enteroids with Leukocyte Component as a Model to Study Host–Pathogen Interactions. Commun. Biol. 2021, 4, 377. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium Persistence and Antibiotic Response in Colorectal Cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Unterer, S.; Busch, K. Acute Hemorrhagic Diarrhea Syndrome in Dogs. Vet. Clin. N. Am. -Small Anim. Pract. 2021, 51, 79–92. [Google Scholar] [CrossRef] [PubMed]
- AlShawaqfeh, M.K.; Wajid, B.; Minamoto, Y.; Markel, M.; Lidbury, J.A.; Steiner, J.M.; Serpedin, E.; Suchodolski, J.S. A Dysbiosis Index to Assess Microbial Changes in Fecal Samples of Dogs with Chronic Inflammatory Enteropathy. FEMS Microbiol. Ecol. 2017, 93, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez-Baeza, Y.; Hyde, E.R.; Suchodolski, J.S.; Knight, R. Dog and Human Inflammatory Bowel Disease Rely on Overlapping yet Distinct Dysbiosis Networks. Nat. Microbiol. 2016, 1, 16177. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting Dysbiosis, Bile-Acid Dysmetabolism and Gut Inflammation in Inflammatory Bowel Diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Kent, A.C.C.; Cross, G.; Taylor, D.R.; Sherwood, R.A.; Watson, P.J. Measurement of Serum 7α-Hydroxy-4-Cholesten-3-One as a Marker of Bile Acid Malabsorption in Dogs with Chronic Diarrhoea: A Pilot Study. Vet. Rec. Open 2016, 3. [Google Scholar] [CrossRef] [PubMed]
- Guard, B.C.; Suchodolski, J.S. Horse Species Symposium: Canine Intestinal Microbiology and Metagenomics: From Phylogeny to Function. J. Anim. Sci. 2016, 94, 2247–2261. [Google Scholar] [CrossRef]
- Weiß, F.; Holthaus, D.; Kraft, M.; Klotz, C.; Schneemann, M.; Schulzke, J.D.; Krug, S.M. Human Duodenal Organoid-Derived Monolayers Serve as a Suitable Barrier Model for Duodenal Tissue. Ann. N. Y. Acad. Sci. 2022. [Google Scholar] [CrossRef]
- Sun, H.; Chow, E.C.Y.; Liu, S.; Du, Y.; Pang, K.S. The Caco-2 Cell Monolayer: Usefulness and Limitations. Expert Opin. Drug Metab. Toxicol. 2008, 4, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Inui, T.; Yokota, J.; Kawakami, K.; Morinaga, G.; Takatani, M.; Hirayama, D.; Nomoto, R.; Ito, K.; Cui, Y.; et al. Monolayer Platform Using Human Biopsy-Derived Duodenal Organoids for Pharmaceutical Research. Mol. Ther. Methods Clin. Dev. 2021, 22, 263–278. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Noguchi, M.; Inoue, Y.; Sato, S.; Shimizu, M.; Kojima, H.; Okabe, T.; Kiyono, H.; Yamauchi, Y.; Sato, R. Organoid-Derived Intestinal Epithelial Cells Are a Suitable Model for Preclinical Toxicology and Pharmacokinetic Studies. iScience 2022, 25, 104542. [Google Scholar] [CrossRef]
- Kozuka, K.; He, Y.; Koo-McCoy, S.; Kumaraswamy, P.; Nie, B.; Shaw, K.; Chan, P.; Leadbetter, M.; He, L.; Lewis, J.G.; et al. Development and Characterization of a Human and Mouse Intestinal Epithelial Cell Monolayer Platform. Stem Cell Rep. 2017, 9, 1976–1990. [Google Scholar] [CrossRef]
- Hoffmann, P.; Schnepel, N.; Langeheine, M.; Künnemann, K.; Grassl, G.A.; Brehm, R.; Seeger, B.; Mazzuoli-Weber, G.; Breves, G. Intestinal Organoid-Based 2D Monolayers Mimic Physiological and Pathophysiological Properties of the Pig Intestine. PLoS ONE 2021, 16, e0256143. [Google Scholar] [CrossRef] [PubMed]
- Ambrosini, Y.M.; Park, Y.; Jergens, A.E.; Shin, W.; Min, S.; Atherly, T.; Borcherding, D.C.; Jang, J.; Allenspach, K.; Mochel, J.P.; et al. Recapitulation of the Accessible Interface of Biopsy-Derived Canine Intestinal Organoids to Study Epithelial-Luminal Interactions. PLoS ONE 2020, 15, e0231423. [Google Scholar] [CrossRef] [PubMed]
- Mayorgas, A.; Dotti, I.; Martínez-Picola, M.; Esteller, M.; Bonet-Rossinyol, Q.; Ricart, E.; Salas, A.; Martínez-Medina, M. A Novel Strategy to Study the Invasive Capability of Adherent-Invasive Escherichia Coli by Using Human Primary Organoid-Derived Epithelial Monolayers. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Miyamoto, K.; Maslowski, K.M.; Ohno, H.; Kanai, T.; Sato, T. Development of a Scalable Coculture System for Gut Anaerobes and Human Colon Epithelium. Gastroenterology 2020, 159, 388–390. [Google Scholar] [CrossRef]
- Bayguinov, P.O.; Oakley, D.M.; Shih, C.C.; Geanon, D.J.; Joens, M.S.; Fitzpatrick, J.A.J. Modern Laser Scanning Confocal Microscopy. Curr. Protoc. Cytom. 2018, 85, e39. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Silva, P.N.; Syed, A.M.; Sindhwani, S.; Rocheleau, J.V.; Chan, W.C.W. Clarifying Intact 3D Tissues on a Microfluidic Chip for High-Throughput Structural Analysis. Proc. Natl. Acad. Sci. USA 2016, 113, 14915–14920. [Google Scholar] [CrossRef] [PubMed]
- van Ineveld, R.L.; Ariese, H.C.R.; Wehrens, E.J.; Dekkers, J.F.; Rios, A.C. Single-Cell Resolution Three-Dimensional Imaging of Intact Organoids. J. Vis. Exp. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- VanDussen, K.L.; Sonnek, N.M.; Stappenbeck, T.S. L-WRN Conditioned Medium for Gastrointestinal Epithelial Stem Cell Culture Shows Replicable Batch-to-Batch Activity Levels across Multiple Research Teams. Stem Cell Res. 2019, 37. [Google Scholar] [CrossRef] [PubMed]
- Janda, C.Y.; Dang, L.T.; You, C.; Chang, J.; de Lau, W.; Zhong, Z.A.; Yan, K.S.; Marecic, O.; Siepe, D.; Li, X.; et al. Surrogate Wnt Agonists That Phenocopy Canonical Wnt and β-Catenin Signalling. Nature 2017, 545, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Ha, A.; de Lau, W.; Yuki, K.; Santos, A.J.M.; You, C.; Geurts, M.H.; Puschhof, J.; Pleguezuelos-Manzano, C.; Peng, W.C.; et al. Next-Generation Surrogate Wnts Support Organoid Growth and Deconvolute Frizzled Pleiotropy In Vivo. Cell Stem Cell 2020, 27, 840–851.e6. [Google Scholar] [CrossRef] [PubMed]
- Luca, V.C.; Miao, Y.; Li, X.; Hollander, M.J.; Kuo, C.J.; Christopher Garcia, K. Surrogate R-Spondins for Tissue-Specific Potentiation of Wnt Signaling. PLoS ONE 2020, 15, e0226928. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793.e6. [Google Scholar] [CrossRef]
- Nantasanti, S.; Spee, B.; Kruitwagen, H.S.; Chen, C.; Geijsen, N.; Oosterhoff, L.A.; van Wolferen, M.E.; Pelaez, N.; Fieten, H.; Wubbolts, R.W.; et al. Disease Modeling and Gene Therapy of Copper Storage Disease in Canine Hepatic Organoids. Stem Cell Rep. 2015, 5, 895–907. [Google Scholar] [CrossRef] [PubMed]
- Aisenbrey, E.A.; Murphy, W.L. Synthetic Alternatives to Matrigel. Nat. Rev. Mater. 2020, 5, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Raphael, B.; Khalil, T.; Workman, V.L.; Smith, A.; Brown, C.P.; Streuli, C.; Saiani, A.; Domingos, M. 3D Cell Bioprinting of Self-Assembling Peptide-Based Hydrogels. Mater. Lett. 2017, 190, 103–106. [Google Scholar] [CrossRef]
- Cherne, M.D.; Sidar, B.; Sebrell, T.A.; Sanchez, H.S.; Heaton, K.; Kassama, F.J.; Roe, M.M.; Gentry, A.B.; Chang, C.B.; Walk, S.T.; et al. A Synthetic Hydrogel, VitroGel® ORGANOID-3, Improves Immune Cell-Epithelial Interactions in a Tissue Chip Co-Culture Model of Human Gastric Organoids and Dendritic Cells. Front Pharmacol. 2021, 12, 2279. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, M.; Malinen, M.M.; Lauren, P.; Lou, Y.R.; Kuisma, S.W.; Kanninen, L.; Lille, M.; Corlu, A.; Guguen-Guillouzo, C.; Ikkala, O.; et al. Nanofibrillar Cellulose Hydrogel Promotes Three-Dimensional Liver Cell Culture. Proc. J. Control. Release 2012, 164, 291–298. [Google Scholar]
- Krüger, M.; Oosterhoff, L.A.; van Wolferen, M.E.; Schiele, S.A.; Walther, A.; Geijsen, N.; de Laporte, L.; van der Laan, L.J.W.; Kock, L.M.; Spee, B. Cellulose Nanofibril Hydrogel Promotes Hepatic Differentiation of Human Liver Organoids. Adv. Healthc. Mater. 2020, 9, 1901658. [Google Scholar] [CrossRef] [PubMed]
- Flaig, I.; Radenković, M.; Najman, S.; Pröhl, A.; Jung, O.; Barbeck, M. In Vivo Analysis of the Biocompatibility and Immune Response of Jellyfish Collagen Scaffolds and Its Suitability for Bone Regeneration. Int. J. Mol. Sci. 2020, 21, 4518. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.J.; Chun, S.M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-Derived Lung Cancer Organoids as in Vitro Cancer Models for Therapeutic Screening. Nat. Commun. 2019, 10, 3991. [Google Scholar] [CrossRef] [PubMed]
- Driehuis, E.; Kretzschmar, K.; Clevers, H. Establishment of Patient-Derived Cancer Organoids for Drug-Screening Applications. Nat. Protoc. 2020, 15, 3380–3409. [Google Scholar] [CrossRef] [PubMed]
- HUB Organoids A Patient in the Lab®. Available online: https://www.huborganoids.nl (accessed on 15 July 2022).
- Sampaziotis, F.; Muraro, D.; Tysoe, O.C.; Sawiak, S.; Beach, T.E.; Godfrey, E.M.; Upponi, S.S.; Brevini, T.; Wesley, B.T.; Garcia-Bernardo, J.; et al. Cholangiocyte Organoids Can Repair Bile Ducts after Transplantation in the Human Liver. Science 2021, 371, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, E.; O’Connor, C.; Gasser, E.; Wei, Z.; Oh, T.G.; Tseng, T.W.; Wang, D.; Cayabyab, F.; Dai, Y.; Yu, R.T.; et al. Immune-Evasive Human Islet-like Organoids Ameliorate Diabetes. Nature 2020, 586, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Meran, L.; Massie, I.; Campinoti, S.; Weston, A.E.; Gaifulina, R.; Tullie, L.; Faull, P.; Orford, M.; Kucharska, A.; Baulies, A.; et al. Engineering Transplantable Jejunal Mucosal Grafts Using Patient-Derived Organoids from Children with Intestinal Failure. Nat. Med. 2020, 26, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Kobayashi, E.; Fujii, M.; Ohta, Y.; Arai, K.; Matano, M.; Ishikawa, K.; Miyamoto, K.; Toshimitsu, K.; Takahashi, S.; et al. An Organoid-Based Organ-Repurposing Approach to Treat Short Bowel Syndrome. Nature 2021, 592, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Koike, H.; Iwasawa, K.; Ouchi, R.; Maezawa, M.; Giesbrecht, K.; Saiki, N.; Ferguson, A.; Kimura, M.; Thompson, W.L.; Wells, J.M.; et al. Modelling Human Hepato-Biliary-Pancreatic Organogenesis from the Foregut–Midgut Boundary. Nature 2019, 574, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, P.; Singh, S.K.; Gulati, M.; Vishwas, S.; Kapoor, B.; Chellappan, D.K.; Anand, K.; Gupta, G.; Jha, N.K.; Gupta, P.K.; et al. Microfluidic Chips: Recent Advances, Critical Strategies in Design, Applications and Future Perspectives. Microfluid Nanofluidics 2021, 25, 99. [Google Scholar] [CrossRef] [PubMed]
- Clapp, N.; Amour, A.; Rowan, W.C.; Candarlioglu, P.L. Organ-on-Chip Applications in Drug Discovery: An End User Perspective. Biochem. Soc. Trans. 2021, 49, 1881–1890. [Google Scholar] [CrossRef] [PubMed]
- Picollet-D’hahan, N.; Zuchowska, A.; Lemeunier, I.; le Gac, S. Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication. Trends Biotechnol. 2021, 39, 788–810. [Google Scholar] [CrossRef]
- Ronaldson-Bouchard, K.; Teles, D.; Yeager, K.; Tavakol, D.N.; Zhao, Y.; Chramiec, A.; Tagore, S.; Summers, M.; Stylianos, S.; Tamargo, M.; et al. A Multi-Organ Chip with Matured Tissue Niches Linked by Vascular Flow. Nat. Biomed. Eng. 2022, 6, 351–371. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Csukovich, G.; Pratscher, B.; Burgener, I.A. The World of Organoids: Gastrointestinal Disease Modelling in the Age of 3R and One Health with Specific Relevance to Dogs and Cats. Animals 2022, 12, 2461. https://doi.org/10.3390/ani12182461
Csukovich G, Pratscher B, Burgener IA. The World of Organoids: Gastrointestinal Disease Modelling in the Age of 3R and One Health with Specific Relevance to Dogs and Cats. Animals. 2022; 12(18):2461. https://doi.org/10.3390/ani12182461
Chicago/Turabian StyleCsukovich, Georg, Barbara Pratscher, and Iwan Anton Burgener. 2022. "The World of Organoids: Gastrointestinal Disease Modelling in the Age of 3R and One Health with Specific Relevance to Dogs and Cats" Animals 12, no. 18: 2461. https://doi.org/10.3390/ani12182461





