Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Samples Collection
2.3. Gene Cloning of Cs-ewsr1-w and Cs-ewsr1-z
2.4. Characterization of Cs-ewsr1-w and Cs-ewsr1-z
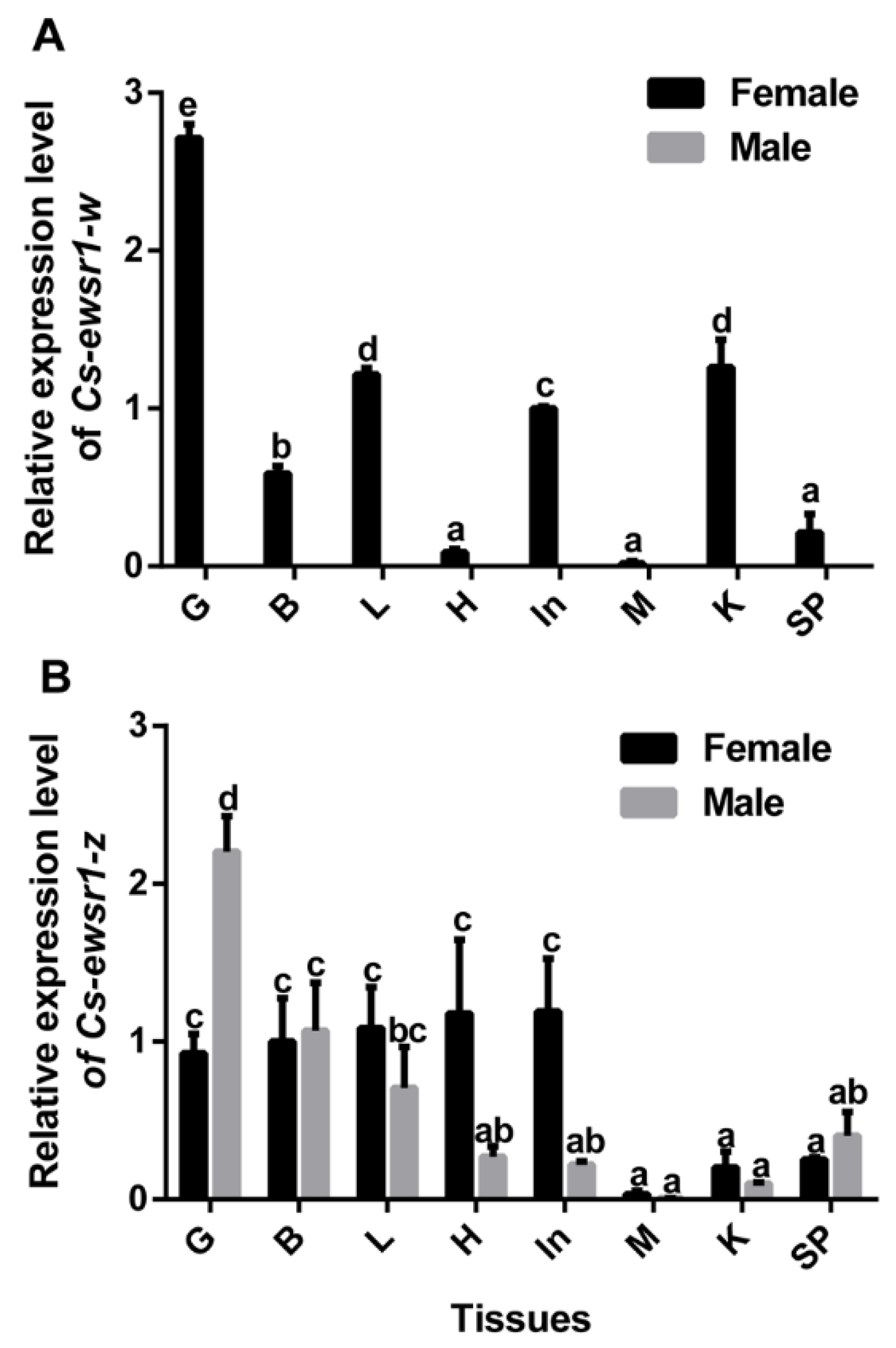
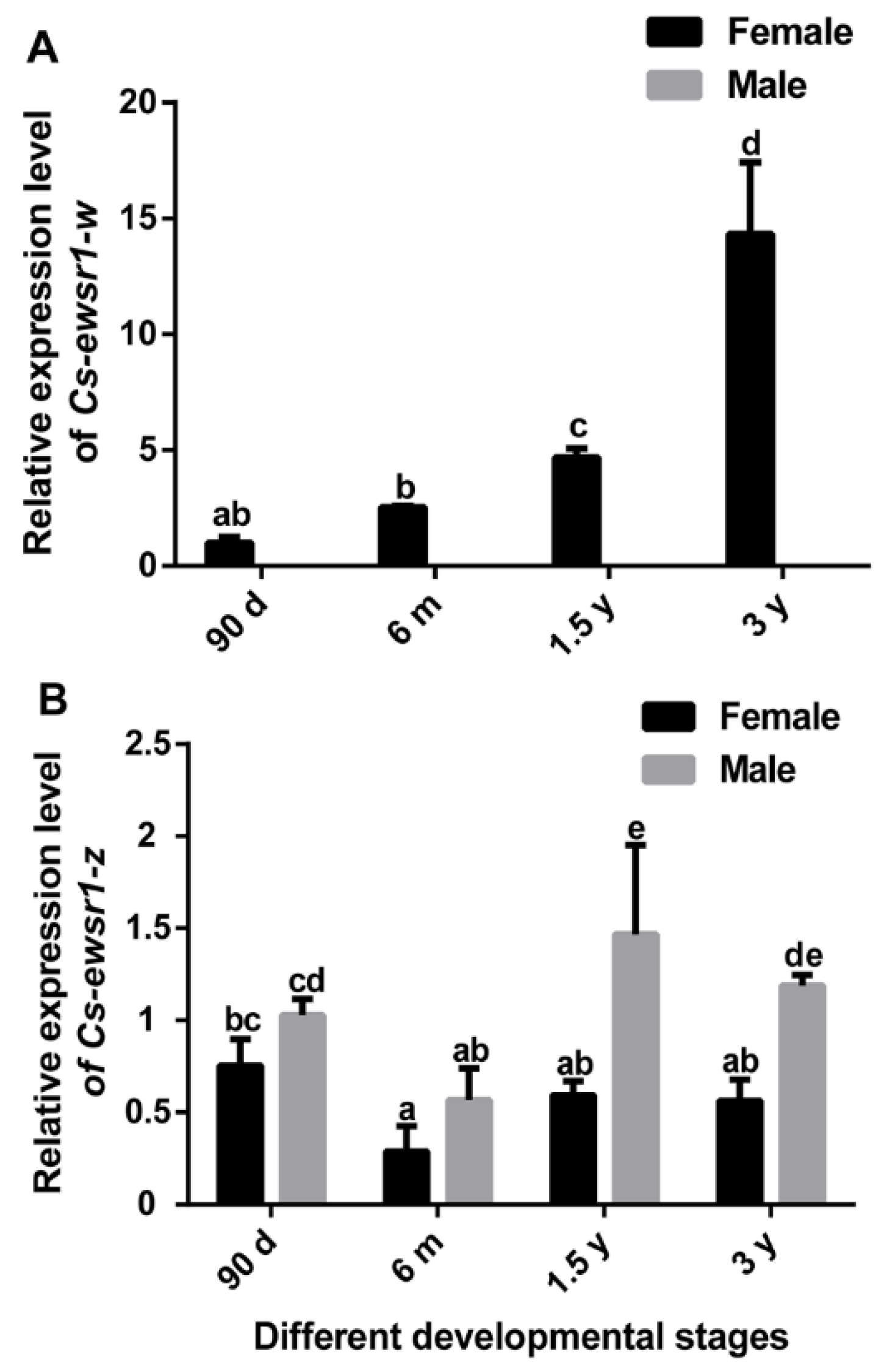
2.5. Gene Expression Patterns of Cs-ewsr1-w and Cs-ewsr1-z in Different Tissues and Stages
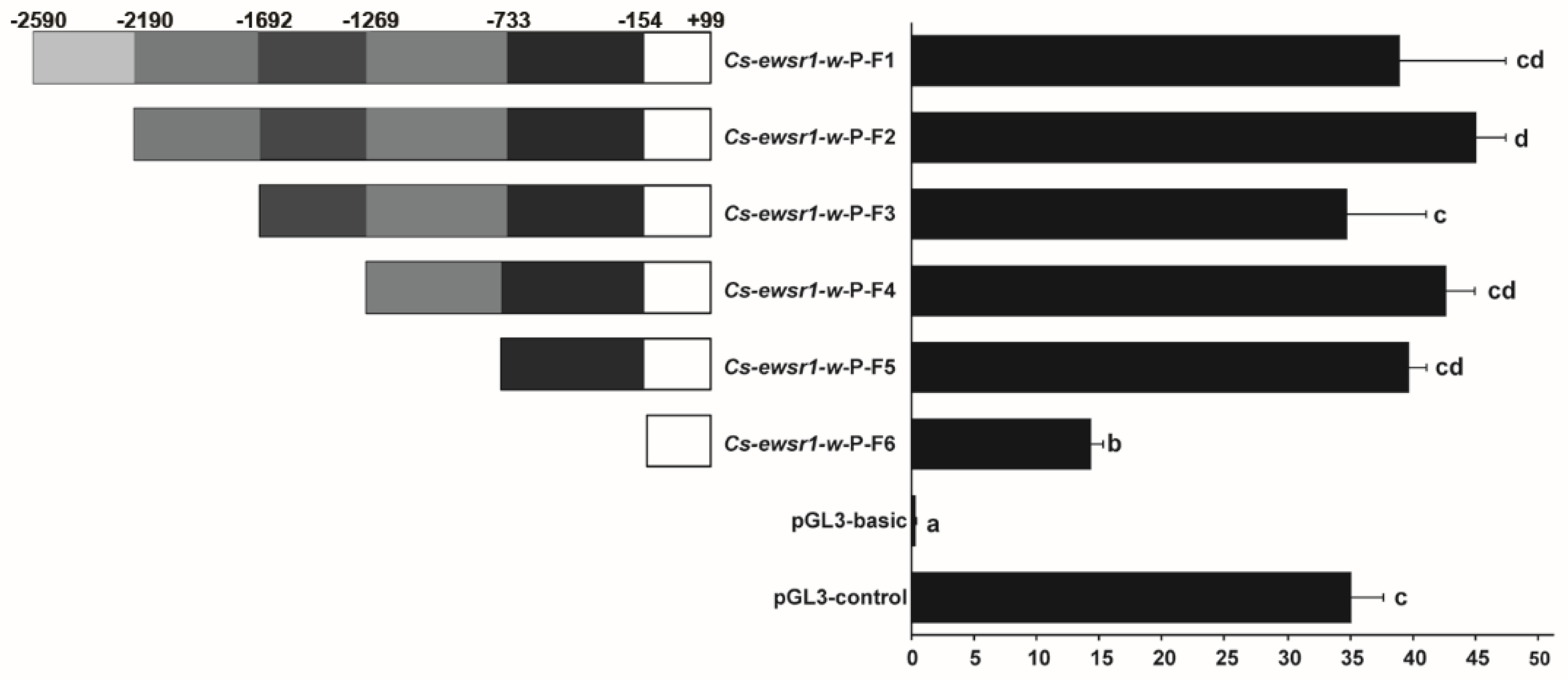
2.6. Promoter Activities Analysis of Cs-ewsr1-w
2.7. The Knockdown Effect of Cs-ewsr1-w siRNA in C. semilaevis Ovarian Cells
3. Results
3.1. Gene Cloning and Characterization of Cs-ewsr1s
3.2. The Expression Patterns of Cs-ewsr1s in Different Tissues and Developmental Stages
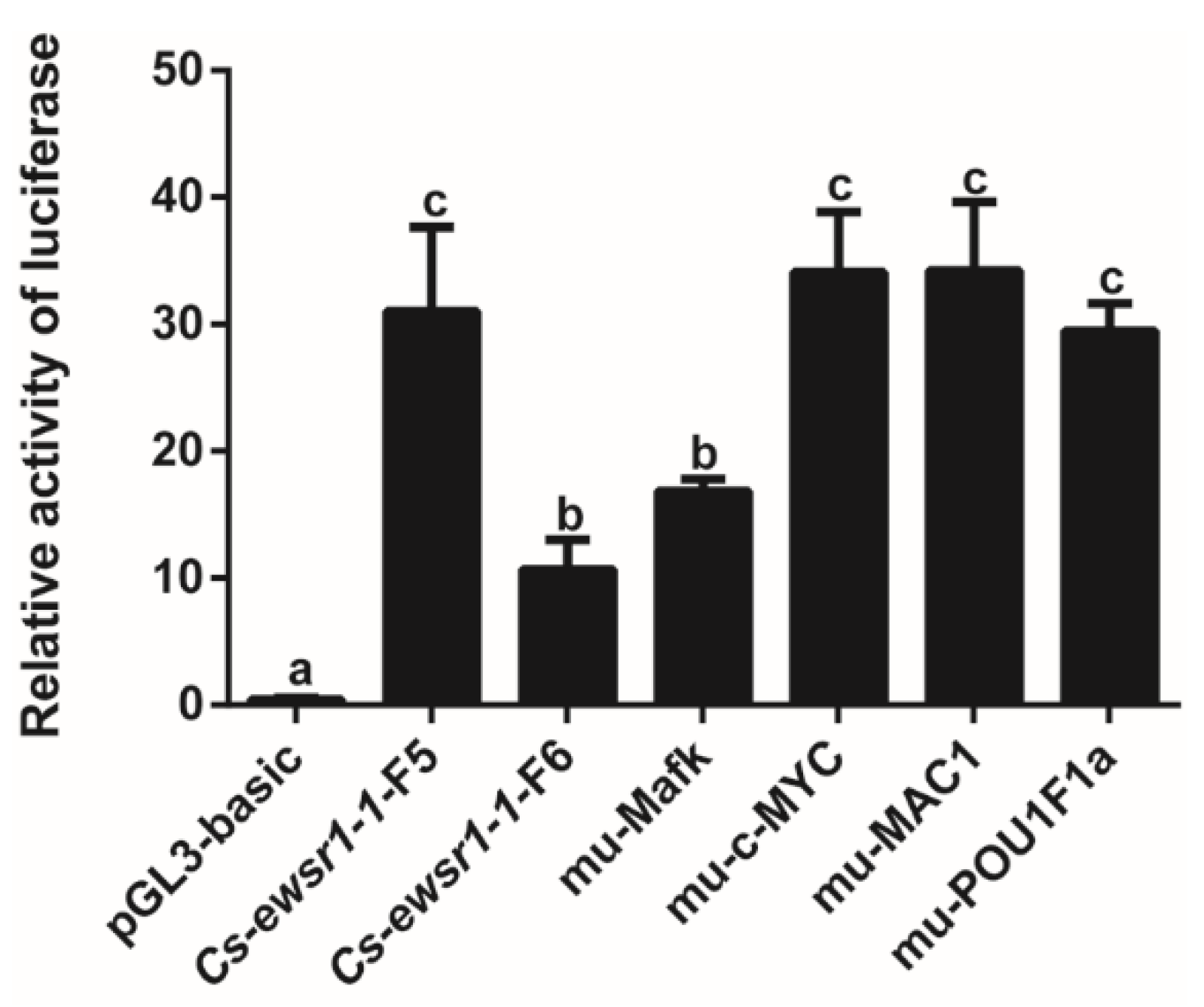
3.3. Promoter Activity of Cs-ewsr1-w Detection and Analysis
3.4. Expression Patterns of Sex-Related Genes in Cs-ewsr1-w Knockdown Ovarian Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Naqvi, S.; Godfrey, A.K.; Hughes, J.F.; Goodheart, M.L.; Mitchell, R.N.; Page, D.C. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 2019, 365, eaaw7317. [Google Scholar] [CrossRef]
- Mei, J.; Gui, J. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef]
- Parker, G.A. The evolution of sexual size dimorphism in fish*. J. Fish Biol. 1992, 41, 1–20. [Google Scholar] [CrossRef]
- Foellmer, M.W.; Moya-Laraño, J. Chater 7. Sexual size dimorphism in spiders: patterns and processes. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 71–81. ISBN 9780191709036. [Google Scholar]
- Lindenfors, P.; Gittleman, J.; Jones, K. Chapter 2. Sexual size dimorphism in mammals. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 16–26. ISBN 9780191709036. [Google Scholar]
- Székely, T.; Lislevand, T.; Figuerola, J. Chapter 3. Sexual size dimorphism in birds. In Sex, Size and Gender Roles: Evolutionary Studies of Sexual Size Dimorphism; Daphne, J.F., Wolf, U.B., Tamás, S., Eds.; Oxford University Press: New York, NY, USA, 2007; pp. 27–37. ISBN 9780191709036. [Google Scholar]
- Sun, Y.; Yu, H.; Zhang, Q.; Qi, J.; Zhong, Q.; Chen, Y.; Li, C. Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010, 155, 316–321. [Google Scholar] [CrossRef]
- Bonduriansky, R.; Chenoweth, S. Intralocus sexual conflict. Trends Ecol. Evol. 2009, 24, 280–288. [Google Scholar] [CrossRef]
- Parsch, J.; Ellegren, H. The evolutionary causes and consequences of sex-biased gene expression. Nat. Rev. Genet. 2013, 14, 83–87. [Google Scholar] [CrossRef]
- Williams, T.M.; Carroll, S.B. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat. Rev. Genet. 2009, 10, 797–804. [Google Scholar] [CrossRef]
- Wang, N.; Wang, R.K.; Wang, R.Q.; Chen, S.L. Transcriptomics analysis revealing candidate networks and genes for the body size sexual dimorphism of Chinese tongue sole (Cynoglossus semilaevis). Funct. Integr. Genom. 2018, 18, 327–339. [Google Scholar] [CrossRef]
- Wang, K.L.; Zhang, H.; Hu, Q.M.; Shao, C.W.; Chen, S.L. Expression and purification of half-smooth tongue sole (Cynoglossus semilaevis) CSDAZL protein. Protein Expr. Purif. 2014, 102, 8–12. [Google Scholar] [CrossRef]
- Wang, N.; Gong, Z.H.; Wang, J.; Xu, W.T.; Yang, Q.; Chen, S.L. Characterization of Chinese tongue sole (Cynoglossus semilaevis) 24-dehydrocholesterol reductase: Expression profile, epigenetic modification, and its knock-down effect. Gen. Comp. Endocrinol. 2021, 312, 113870. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Li, X.H.; Shi, R.; Cheng, P.; Wang, N.; Chen, S.L. The female-biased expression, transcriptional regulation and knock-down effect of insulin-like growth factor binding protein 7 in Chinese tongue sole, Cynoglossus semilaevis. Aquaculture 2022, 551, 737956. [Google Scholar] [CrossRef]
- Xu, W.T.; Cui, Z.K.; Wang, N.; Zhang, M.Q.; Wang, J.; Xu, X.T.; Liu, Y.; Chen, S.L. Transcriptomic analysis revealed gene expression profiles during the sex differentiation of Chinese tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 40, 100919. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nguyen, P.T.; Shim, H.S.; Hyeon, S.J.; Im, H.; Choi, M.H.; Chung, S.; Kowall, N.W.; Lee, S.B.; Ryu, H. EWSR1, a multifunctional protein, regulates cellular function and aging via genetic and epigenetic pathways. Biochim. Biophys. Acta-(BBA) Mol. Basis Dis. 2019, 1865, 1938–1945. [Google Scholar] [CrossRef] [PubMed]
- Rossow, K.L.; Janknecht, R. The Ewing’s sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001, 61, 2690–2695. [Google Scholar]
- Erkizan, H.; Uversky, V.; Toretsky, J. Oncogenic Partnerships: EWS-FLI1 Protein Interactions Initiate Key Pathways of Ewing’s Sarcoma. Clin. Cancer Res. 2010, 16, 4077–4083. [Google Scholar] [CrossRef] [PubMed]
- Morohoshi, F.; Ootsuka, Y.; Arai, K.; Ichikawa, H.; Mitani, S.; Munakata, N.; Ohki, M. Genomic structure of the human RBP56/hTAF(II)68 and FUS/TLS genes. Gene 1998, 221, 191–198. [Google Scholar] [CrossRef]
- Bertolotti, A.; Melot, T.; Acker, J.; Vigneron, M.; Delattre, O.; Tora, L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: Interactions between two members of the tet family, EWS and HTAF(II)68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998, 18, 1489–1497. [Google Scholar] [CrossRef]
- Azuma, M.; Embree, L.J.; Sabaawy, H.; Hickstein, D.D. Ewing sarcoma protein ewsr1 maintains mitotic integrity and proneural cell survival in the zebrafish embryo. PLoS ONE 2007, 2, e979. [Google Scholar] [CrossRef]
- Li, H.; Watford, W.; Li, C.; Parmelee, A.; Bryant, M.A.; Deng, C.; O’Shea, J.; Lee, S.B. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J. Clin. Investig. 2007, 117, 1314–1323. [Google Scholar] [CrossRef]
- Araya, N.; Hirota, K.; Shimamoto, Y.; Miyagishi, M.; Yoshida, E.; Ishida, J.; Kaneko, S.; Kaneko, M.; Nakajima, T.; Fukamizu, A. Cooperative interaction of EWS with CREB-binding protein selectively activates hepatocyte nuclear factor 4-mediated transcription. J. Biol. Chem. 2003, 278, 5427–5432. [Google Scholar] [CrossRef]
- Lee, J.; Rhee, B.K.; Bae, G.Y.; Han, Y.M.; Kim, J. Stimulation of Oct-4 activity by Ewing’s sarcoma protein. Stem Cells 2005, 23, 738–751. [Google Scholar] [CrossRef] [PubMed]
- Kovar, H. Dr. Jekyll and Mr. Hyde: The Two Faces of the FUS/EWS/TAF15 Protein Family. Sarcoma 2011, 2011, 837474. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Han, K.; Chen, S.; Cai, M.; Wang, Y.; Zhang, Z. Molecular cloning and expression of Octamer-binding transcription factor (Oct4) in the large yellow croaker, Larimichthys crocea. Gene Expr. Patterns 2018, 27, 16–30. [Google Scholar] [CrossRef]
- Lanier, J.; Quina, L.A.; Eng, S.R.; Cox, E.; Turner, E.E. Brn3a target gene recognition in embryonic sensory neurons. Dev. Biol. 2007, 302, 703–716. [Google Scholar] [CrossRef]
- Tian, H.; Petkov, P.M. Mouse EWSR1 is crucial for spermatid post-meiotic transcription and spermiogenesis. Development 2021, 148, dev199414. [Google Scholar] [CrossRef]
- Sun, Y.X.; Zhu, Y.; Cheng, P.; Zhang, M.Q.; Wang, N.; Cui, Z.K.; Wei, M.; Xu, W.T. A Z-Linked E3 Ubiquitin Ligase Cs-rchy1 Is Involved in Gametogenesis in Chinese Tongue Sole, Cynoglossus semilaevis. Animals 2021, 11, 3265. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Wang, T.Z.; Wang, N.; Liu, X.F.; Sha, Z.X.; Chen, S.L. Establishment and characterization of an ovarian cell line from half-smooth tongue sole Cynoglossus semilaevis. J. Fish Biol. 2015, 86, 46–59. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, S.L.; Gao, F.T.; Meng, L.; Hu, Q.M.; Song, W.; Shao, C.W.; Lv, W.Q. SCAR-transformation of sex-specific SSR marker and its application in half-smooth tongue sole (Cynoglossus semiliaevis). Agric. Biotechnol. 2014, 6, 787–792. [Google Scholar] [CrossRef]
- Chen, S.L.; Zhang, G.J.; Shao, C.W.; Huang, Q.F.; Liu, G.; Zhang, P.; Song, W.T.; An, N.; Chalopin, D.; Volff, J.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Leturnic, I.; Khedkar, S.; Bork, P. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis acorss computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Messeguer, X.; Escudero, R.; Farré, D.; Nuñez, O.; Martínez, J.; Albà, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Lemma, R.B.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Pérez, N.M.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef]
- Paronetto, M.P. Ewing sarcoma protein: A key player in human cancer. Int. J. Cell Biol. 2013, 2013, 642853. [Google Scholar] [CrossRef]
- Bertolotti, A.; Lutz, Y.; Heard, D.J.; Chambon, P.; Tora, L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996, 15, 5022–5031. [Google Scholar] [CrossRef]
- Uhlenhaut, N.H.; Treier, M. Foxl2 function in ovarian development. Mol. Genet. Metab. 2006, 88, 225–234. [Google Scholar] [CrossRef]
- Okada, H.; Hagihara, S.; Yamashita, K.; Ijiri, S.; Adachi, S. Expression pattern of foxl2 and dmrt1 in gonad of Amur sturgeon Acipenser schrenckii in relation to sex differentiation. Aquaculture 2017, 479, 712–720. [Google Scholar] [CrossRef]
- Alam, M.A.; Kobayashi, Y.; Horiguchi, R.; Hirai, T.; Nakamura, M. Molecular cloning and quantitative expression of sexually dimorphic markers Dmrt1 and Foxl2 during female-to-male sex change in Epinephelus merra. Gen. Comp. Endocrinol. 2008, 157, 75–85. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Yamaguchi, S.; Hirai, T.; Kitano, T. Follicle-stimulating hormone signaling and Foxl2 are involved in transcriptional regulation of aromatase gene during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2007, 359, 935–940. [Google Scholar] [CrossRef]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.L.; Chen, S.L.; Ji, X.S.; Shao, C.W. Molecular cloning, characterization and expression analysis of Sox9a and Foxl2 genes in half-smooth tongue sole (Cynoglossus semilaevis). Acta Oceanol. Sin. 2011, 30, 68–77. [Google Scholar] [CrossRef]
- Chiang, E.F.; Pai, C.I.; Wyatt, M.; Yan, Y.L.; Postlethwait, J.; Chung, B. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001, 231, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Cui, J.; Yang, G.; Gong, Q.; Gu, Q. Expression detection of DMRTs and two sox9 genes in Takifugu rubripes (Tetraodontidae, Vertebrata). J. Ocean Univ. China 2007, 6, 182–186. [Google Scholar] [CrossRef]
- Nakamura, S.; Aoki, Y.; Saito, D.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Tanaka, M. Sox9b/sox9a2-EGFP transgenic medaka reveals the morphological reorganization of the gonads and a common precursor of both the female and male supporting cells. Mol. Reprod. Dev. 2008, 75, 472–476. [Google Scholar] [CrossRef]
- Nakamura, S.; Watakabe, I.; Nishimura, T.; Toyoda, A.; Taniguchi, Y.; Tanaka, M. Analysis of Medaka sox9 Orthologue Reveals a Conserved Role in Germ Cell Maintenance. PLoS ONE 2012, 7, e29982. [Google Scholar] [CrossRef]
- Li, X.; Yu, H.; Wang, Y.; Liu, X.; Liu, Y.; Qu, J.; Wang, X. Roles of Two Sox9 Genes during Gonadal Development in Japanese Flounder: Sex Differentiation, Spermatogenesis and Gonadal Function Maintenance. Int. J. Mol. Sci. 2018, 19, 512. [Google Scholar] [CrossRef]
- Sun, Y.; Zhang, M.Q.; Cheng, P.; Gong, Z.H.; Li, X.H.; Wang, N.; Wei, M.; Xu, X.W.; Xu, W.T. pitpbeta_w Encoding Phosphatidylinositol Transfer Protein Is Involved in Female Differentiation of Chinese Tongue Sole, Cynoglossus semilaevis. Front. Genet. 2022, 13, 861763. [Google Scholar] [CrossRef]
- Xiaohuan, H.; Yang, Z.; Linyan, L.; Zhenhua, F.; Linyan, Z.; Zhijian, W.; Ling, W.; Deshou, W.; Jing, W. Characterization of the POU5F1 Homologue in Nile Tilapia: From Expression Pattern to Biological Activity. Stem Cells Dev. 2016, 25, 1386–1395. [Google Scholar] [CrossRef]
- Zhong, C.; Liu, M.; Tao, Y.; Wu, X.; Yang, Y.; Wang, T.; Meng, Z.; Xu, H.; Liu, X. Pou5f1 and Nanog Are Reliable Germ Cell-Specific Genes in Gonad of a Protogynous Hermaphroditic Fish, Orange-Spotted Grouper (Epinephelus coioides). Genes 2021, 13, 79. [Google Scholar] [CrossRef]
- Gao, J.; Wang, X.; Zhang, Q. Evolutionary Conservation of pou5f3 Genomic Organization and Its Dynamic Distribution during Embryogenesis and in Adult Gonads in Japanese Flounder Paralichthys olivaceus. Int. J. Mol. Sci. 2017, 18, 231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shijun, L.; Khan, R.; Raza, S.H.A.; Jieyun, H.; Chugang, M.; Kaster, N.; Gong, C.; Chunping, Z.; Schreurs, N.M.; Linsen, Z. Function and characterization of the promoter region of perilipin 1 (PLIN1): Roles of E2F1, PLAG1, C/EBPβ, and SMAD3 in bovine adipocytes. Genomics 2020, 112, 2400–2409. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Richmond, T. Eukaryotic transcription factors. Curr. Opin. Struct. Biol. 1998, 8, 41–48. [Google Scholar] [CrossRef]
- de Vooght, K.M.; van Solinge, W.W. Gene promoter analysis in molecular diagnostics: Do or don’t? Expert Rev. Mol. Diagn. 2009, 9, 403–405. [Google Scholar] [CrossRef]
- Yamazaki, H.; Katsuoka, F.; Motohashi, H.; Engel, J.D.; Yamamoto, M. Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol. Cell. Biol. 2012, 32, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.B.; Solovieva, V.; Blank, V. The small MAF transcription factors MAFF, MAFG and MAFK: Current knowledge and perspectives. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2012, 1823, 1841–1846. [Google Scholar] [CrossRef] [PubMed] [Green Version]

 ” represents RNA recognition motif, “
” represents RNA recognition motif, “ ” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
 ” represents RNA recognition motif, “
” represents RNA recognition motif, “ ” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.
” respresents Ran binding protein zinc finger domain, “▲” represents Ewsr1 proteins in C. semilaevis.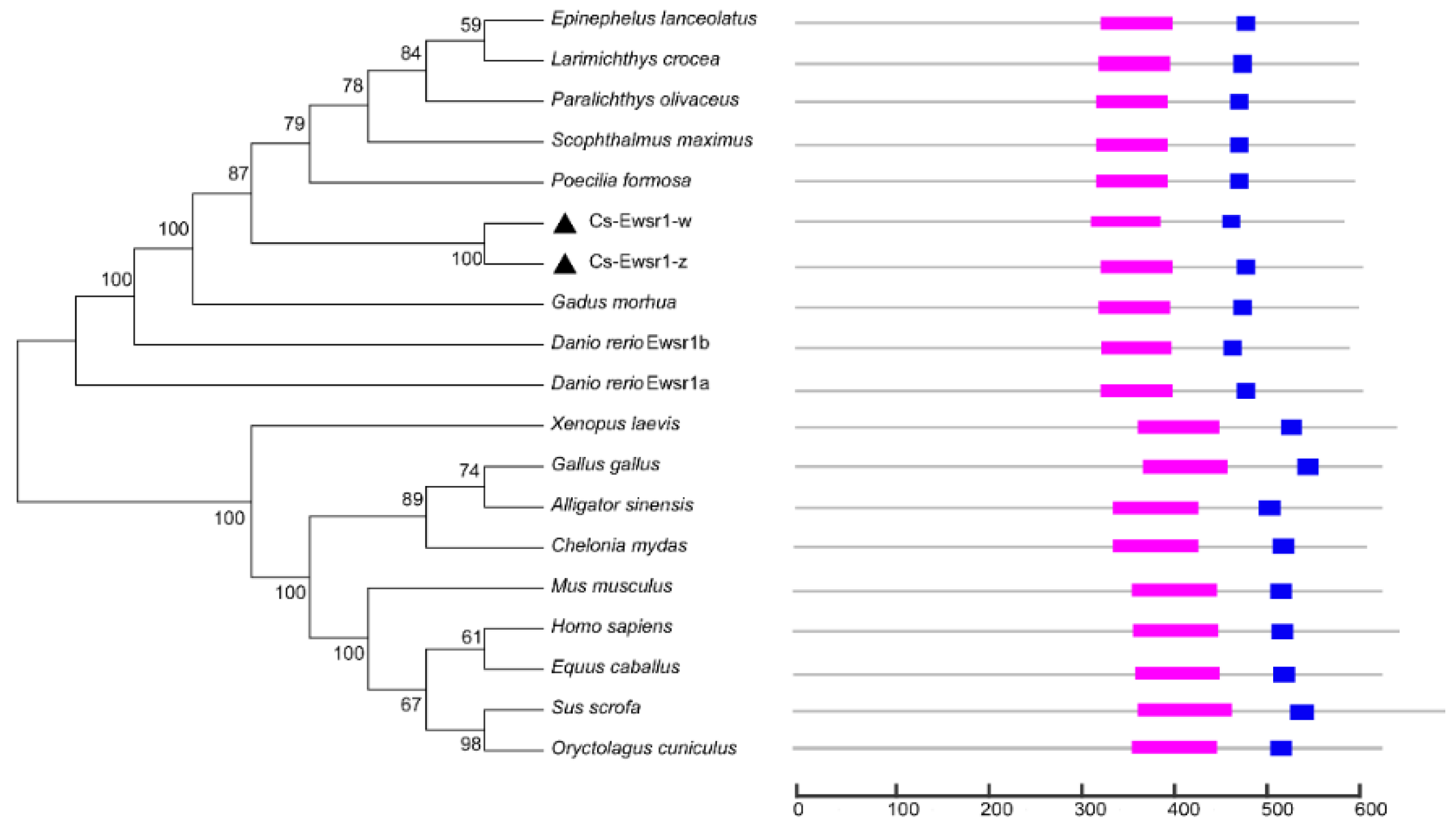


| Symbol | Information | Sequences |
|---|---|---|
| Cs-ewsr1-w-F | CDS cloning | TTCAGTCGTTTAACTCGGAAGT |
| Cs-ewsr1-w-R | CDS cloning | TCACAAGTGGGAAAGTCACCCT |
| Cs-ewsr1-w-5′-1 | 5′-UTR | CTGGAGCAGCAGCAGCTGGAGTACT |
| Cs-ewsr1-w-5′-2 | 5′-UTR | TAACCCTGCTGGGCGTTGGCTTGA |
| Cs-ewsr1-w-3′-1 | 3′-UTR | AATGAGAGGCAGCATGCCAATAAGA |
| Cs-ewsr1-w-3′-2 | 3′-UTR | GGTCGGCGGTCCTCCATTCCCCCCT |
| Cs-ewsr1-w-RT-F | qPCR | GCGGGCCCCCCATGGACC |
| Cs-ewsr1-w-RT-R | qPCR | CAAATTTCACAAGTGGGA |
| Cs-ewsr1-w-P-F1 | promoter | AGATCTGCGATCTAAGTAAGCTGACGCTGGCATGTATGTT |
| Cs-ewsr1-w-P-F2 | promoter | AGATCTGCGATCTAAGTAAGCTGAGGACCACAACGACCCA |
| Cs-ewsr1-w-P-F3 | promoter | AGATCTGCGATCTAAGTAAGCTTGCGAACAAATCACTGCG |
| Cs-ewsr1-w-P-F4 | promoter | AGATCTGCGATCTAAGTAAGCTAGGTGTATCCTAAACAGAAA |
| Cs-ewsr1-w-P-F5 | promoter | AGATCTGCGATCTAAGTAAGCTATCGGATCGGAAAGGAAA |
| Cs-ewsr1-w-P-F6 | promoter | AGATCTGCGATCTAAGTAAGCTCGGTCTTCCCATCACTAA |
| Cs-ewsr1-w-P-R | promoter | CAACAGTACCGGAATGCCAAGCTTTCCGAGTTAAACGACTGAAGA |
| mu-Mafk-F | TF binding site mutation | CAAACCATGTTGCCTCGTACGCTAAGTAAGCCCTACT |
| mu-Mafk-R | TF binding site mutation | CAAACCATGTTGCCTTCAGTATTAAGTAAGCCCTACT |
| mu-c-Myc-F | TF binding site mutation | ATTAGTTTCATTTGTAGACGAACAGAGAGCTCGGGT |
| mu-c-Myc-R | TF binding site mutation | ACCCGAGCTCTCTGTTCGTCTACAAATGAAACTAAT |
| mu-MAC1-F | TF binding site mutation | AGAGCTCGGGTCTCTACGATGATCATTCATCTTCACA |
| mu-MAC1-R | TF binding site mutation | TGTGAAGATGAATGATCATCGTAGAGACCCGAGCTCT |
| mu-POU1F1a-F | TF binding site mutation | AGTAAGCCCTACTCTCGGCTGTGTCAGAGAACCGC |
| mu-POU1F1a-R | TF binding site mutation | GCGGTTCTCTGACACAGCCGAGAGTAGGGCTTACT |
| Cs-ewsr1-w-siRNA | siRNA | CGUUUAACUCGGAAGUUGUGA |
| sox9b-F | qPCR | AAGAACCACACAGATCAAGACAGA |
| sox9b-R | qPCR | TAGTCATACTGTGCTCTGGTGATG |
| foxl2-F | qPCR | GAGGAAGGGCAACTACTGGA |
| foxl2-R | qPCR | CAGCGACCAGGAGTTGTTCA |
| pou5f1-F | qPCR | CCATCTGCCGCTTTGAGG |
| pou5f1-R | qPCR | CCTGGGTGTTGGGTTTGG |
| Cs-ewsr1-z-F | CDS cloning | ATGGCGTCGACACAGGATTACAGCT |
| Cs-ewsr1-z-R | CDS cloning | TTAGTAAGGTCTGTCTCGGCGCTCC |
| Cs-ewsr1-z-5′-1 | 5′-UTR | CAGCAGCTGGAGTACTGTCATATCC |
| Cs-ewsr1-z-5′-2 | 5′-UTR | CCCCTGCTGGGCGCTGGTCTGG |
| Cs-ewsr1-z-3′-1 | 3′-UTR | GATGAGAGGTGGCATGCCAATGAGA |
| Cs-ewsr1-z-3′-2 | 3′-UTR | AGGCGGAGGTCCTCCATTTCCCCCT |
| Cs-ewsr1-z-RT-F | qPCR | GGATATGACAGTACTCCAGCT |
| Cs-ewsr1-z-RT-R | qPCR | TCCTGCAGGCTGGCTATAGCTAC |
| sex-F | sex identification | CCTAAATGATGGATGTAGATTCTGTC |
| sex-R | sex identification | GATCCAGAGAAAATAAACCCAGG |
| Species | Accession No. |
|---|---|
| Larimichthys crocea | XP_019130992.1 |
| Paralichthys olivaceus | XP_019956591.1 |
| Poecilia formosa | XP_007559175.1 |
| Gadus morhua | XP_030215256.1 |
| Scophthalmus maximus | XP_035495384.1 |
| Danio rerio-Ewsr1a | XP_021334784.1 |
| Danio rerio-Ewsr1b | NP_997795.1 |
| Epinephelus lanceolatus | XP_033475149.1 |
| Alligator sinensis | XP_025048938.1 |
| Homo sapiens | XP_011528297.1 |
| Gallus gallus | XP_015150339.2 |
| Mus musculus | NP_001269990.1 |
| Chelonia mydas | XP_037735001.1 |
| Sus scrofa | XP_020927659.1 |
| Equus caballus | XP_023502669.1 |
| Xenopus laevis | XP_018095208.1 |
| Oryctolagus cuniculus | XP_017206143.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, P.; Chen, Z.; Xu, W.; Wang, N.; Yang, Q.; Shi, R.; Li, X.; Cui, Z.; Cheng, J.; Chen, S. Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis. Animals 2022, 12, 2503. https://doi.org/10.3390/ani12192503
Cheng P, Chen Z, Xu W, Wang N, Yang Q, Shi R, Li X, Cui Z, Cheng J, Chen S. Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis. Animals. 2022; 12(19):2503. https://doi.org/10.3390/ani12192503
Chicago/Turabian StyleCheng, Peng, Zhangfan Chen, Wenteng Xu, Na Wang, Qian Yang, Rui Shi, Xihong Li, Zhongkai Cui, Jiayu Cheng, and Songlin Chen. 2022. "Potential Involvement of ewsr1-w Gene in Ovarian Development of Chinese Tongue Sole, Cynoglossus semilaevis" Animals 12, no. 19: 2503. https://doi.org/10.3390/ani12192503





