The European Pine Marten Martes martes (Linnaeus, 1758) Is Autochthonous in Sicily and Constitutes a Well-Characterised Major Phylogroup within the Species (Carnivora, Mustelidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
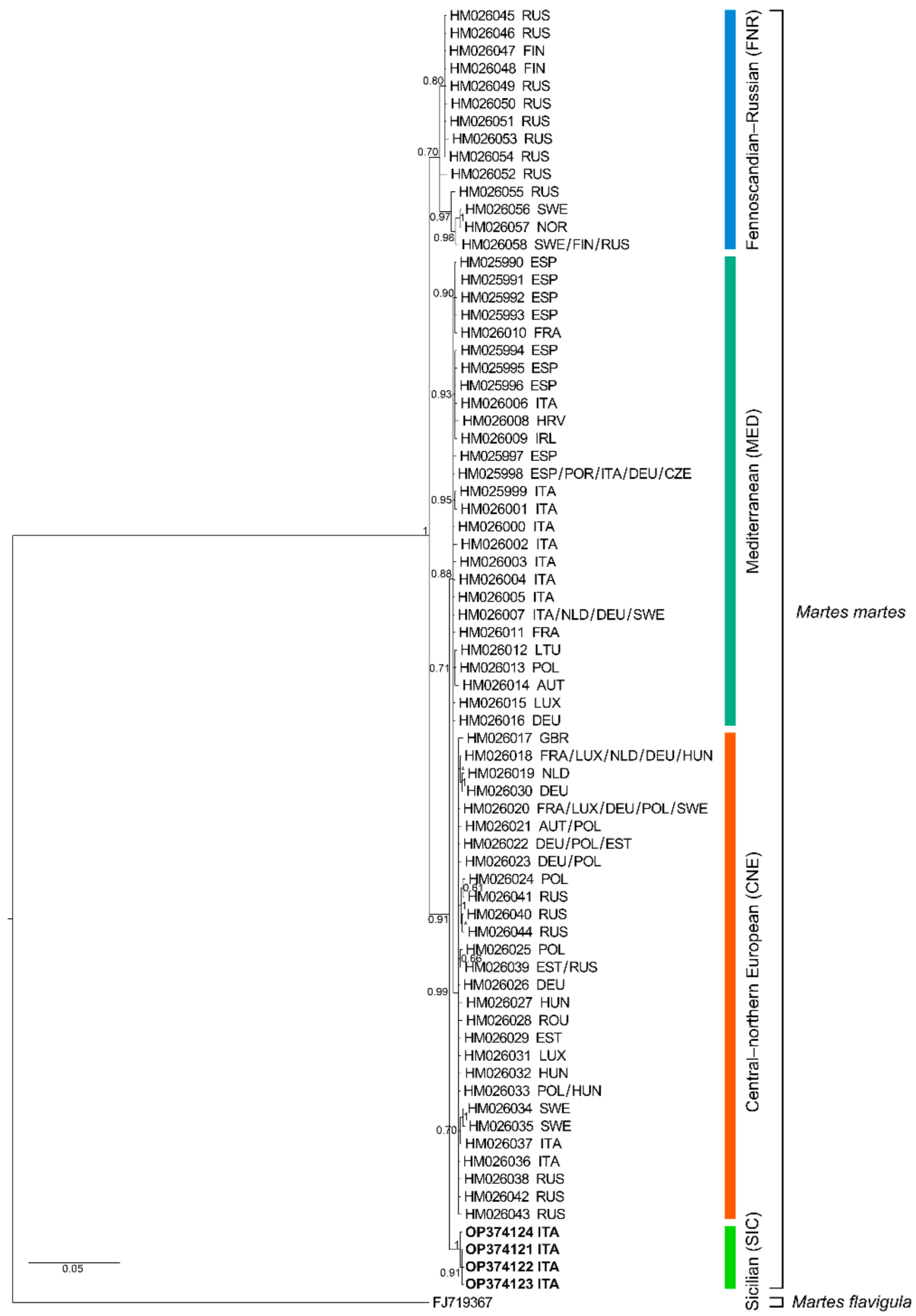
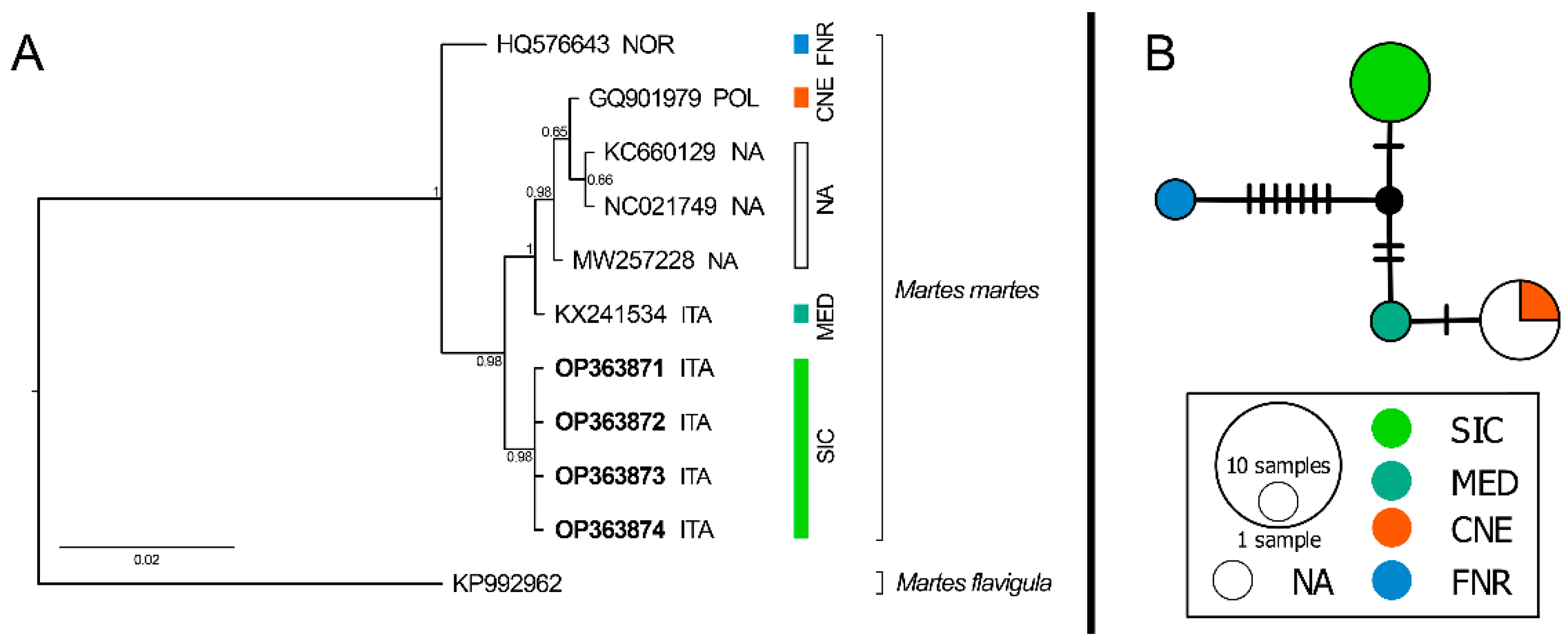
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- La Mantia, T.; Cannella, Z. Presenza storica dei grossi mammiferi in Sicilia. In Atlante della Biodiversità della Sicilia: Vertebrati terrestri. Studi e Ricerche 6; Arpa Sicilia: Palermo, Italy, 2008; pp. 87–106. ISBN 9788895813028. [Google Scholar]
- AUTORI VARI. Atlante della Biodiversità della Sicilia: Vertebrati terrestri. Studi e Ricerche 6; Arpa Sicilia: Palermo, Italy, 2008; ISBN 9788895813028. [Google Scholar]
- da Silveira Bueno, R.; Falcone, S.; La Mantia, T.; Librera, M.; Lo Duca, R.; Seminara, S.; Siracusa, M.; Spinnato, S.; Surdo, S. Update of the distribution and habitat use of the wildcat, pine marten and weasel in Sicily. In Life on Islands. 1. Biodiversity in Sicily and Surrounding Islands. Studies Dedicated to Bruno Massa; La Mantia, T., Badalamenti, E., Carapezza, A., Lo Cascio, P., Troia, A., Eds.; Edizioni Danaus: Palermo, Italy, 2020; pp. 125–150. [Google Scholar]
- Boitani, L.; Lovari, S.; Vigna Taglianti, A. Fauna d’Italia. Mammalia III. Carnivora-Artiodactyla. Vol. XXXVIII; Boitani, L., Lovari, S., Vigna Taglianti, A., Eds.; de Il Sole 24 ORE S.p.A: Bologna, Italy, 2003. [Google Scholar]
- Proulx, G.; Aubry, K.B.; Birks, J. World distribution and status of the genus Martes in 2000. In Martens and Fishers (Martes) in Human-Altered Environments: An International Perspective; Harrison, D.J., Fuller, A.K., Proulx, G., Eds.; Springer: New York, NY, USA, 2004; pp. 77–98. [Google Scholar]
- Herrero, J.; Kranz, A.; Skumatov, D.; Abramov, A.V.; Maran, T.; Monakhov, V.G. Martes martes. IUCN Red List Threat. Species 2016, 8235, e.T12848A45199169. [Google Scholar]
- Burgin, C.J.; Wilson, D.E.; Mittermeier, R.A.; Rylands, A.B.; Lacher, T.E.; Sechrest, W. Illustrated Checklist of the Mammals of the World. Volume 2: Eulipotyphla to Carnivora; Lynx Edicions: Barcelona, Spain, 2020. [Google Scholar]
- Alcover, J.A.; Delibes, M.; Gosálbez, J.; Nadal, J. Martes martes Linnaeus, 1758 a les Balears. Misc. Zool. 1986, 10, 323–333. [Google Scholar]
- Yalden, D.W. The History of British Mammals; T & AD Poyser: London, UK, 1999. [Google Scholar]
- Genovesi, P. Martes Martes. In IUCN 2011. IUCN Red List of Threatened Species. Version 2011.2. Available online: http://www.iucn.it/scheda.php?id=-1546631900 (accessed on 20 August 2022).
- Masseti, M. Uomini e (non solo) Topi. Gli Animali Domestici e la Fauna Antropocora; University Press/Università di Firenze: Florence, Italy, 2008. [Google Scholar]
- Valenzuela, A.; Alcover, J.A. The chronology of the introduction of two species of Martes (Carnivora, Mustelidae) on the Western Mediterranean Islands: First direct radiocarbon evidence. Biol. Invasions 2015, 17, 3093–3100. [Google Scholar] [CrossRef]
- Corbet, G.B. The Mammals of Palearctic Region: A Taxonomic Review; British Museum: London, UK, 1978. [Google Scholar]
- Davison, A.; Birks, J.D.S.; Brookes, R.C.; Messenger, J.E.; Griffiths, H.I. Mitochondrial phylogeography and population history of pine martens Martes martes compared with polecats Mustela putorius. Mol. Ecol. 2001, 10, 2479–2488. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, A.; Madeira, M.J.; Randi, E.; Abramov, A.V.; Davoli, F.; Gómez-Moliner, B.J. Phylogeography of the forest-dwelling European pine marten (Martes martes): New insights into cryptic northern glacial refugia. Biol. J. Linn. Soc. 2013, 109, 1–18. [Google Scholar] [CrossRef]
- Pertoldi, C.; Elschot, K.; Ruiz-González, A.; van de Zande, L.; Zalewski, A.; Muñoz, J.; Madsen, A.B.; Loeschcke, V.; de Groot, A.; Bijlsma, R. Genetic variability of central–western European pine marten (Martes martes) populations. Acta Theriol. 2014, 59, 503–510. [Google Scholar] [CrossRef]
- Colli, L.; Cannas, R.; Deiana, A.M.; Tagliavini, J. Microsatellite variability of Sardinian pine martens, Martes martes. Zool. Sci. 2011, 28, 580–586. [Google Scholar] [CrossRef]
- Genovesi, P.; De Marinis, A.M. Martes martes (Linnaeus, 1758). In Fauna d’Italia. Mammalia III. Carnivora-Artiodactyla. Vol. XXXVIII; Boitani, L., Lovari, S., Vigna Taglianti, A., Eds.; de Il Sole 24 ORE S.p.A: Milano, Italy, 2003; pp. 103–104. [Google Scholar]
- Pertoldi, C.; Breyne, P.; Cabria, M.T.; Halfmaerten, D.; Jansman, H.A.H.; Van Den Berge, K.; Madsen, A.B.; Loeschcke, V. Genetic structure of the European polecat (Mustela putorius) and its implication for conservation strategies. J. Zool. 2006, 270, 102–115. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit 1 from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Richterich, P. Estimation of errors in “Raw” DNA sequences: A validation study. Genome Res. 1998, 8, 251–259. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Dubey, S.; Koyasu, K.; Parapanov, R.; Ribi, M.; Hutterer, R.; Vogel, P. Molecular phylogenetics reveals Messinian, Pliocene, and Pleistocene colonizations of islands by North African shrews. Mol. Phylogenet. Evol. 2008, 47, 877–882. [Google Scholar] [CrossRef]
- Seddon, J.M.; Santucci, F.; Reeve, N.J.; Hewitt, G.M. DNA footprints of European hedgehogs, Erinaceus europaeus and E. concolor: Pleistocene refugia, postglacial expansion and colonization routes. Mol. Ecol. 2001, 10, 2187–2198. [Google Scholar] [CrossRef]
- Michaux, J.R.; Magnanou, E.; Paradis, E.; Nieberding, C.; Libois, R. Mitochondrial phylogeography of the woodmouse (Apodemus sylvaticus) in the Western Palearctic region. Mol. Ecol. 2003, 12, 685–697. [Google Scholar] [CrossRef]
- Bezerra, A.M.R.; Annesi, F.; Aloise, G.; Amori, G.; Giustini, L.; Castiglia, R. Integrative taxonomy of the Italian pine voles, Microtus savii group (Cricetidae, Arvicolinae). Zool. Scr. 2015, 45, 225–236. [Google Scholar] [CrossRef]
- Hürner, H.; Krystufek, B.; Sar, M.; Ribas, A.; Ruch, T.; Sommer, R.; Ivashkina, V.; Michaux, J.R. Mitochondrial phylogeography of the edible dormouse (Glis glis) in the western palearctic region. J. Mammal. 2010, 91, 233–242. [Google Scholar] [CrossRef]
- Mattucci, F.; Oliveira, R.; Lyons, L.A.; Alves, P.C.; Randi, E. European wildcat populations are subdivided into five main biogeographic groups: Consequences of Pleistocene climate changes or recent anthropogenic fragmentation? Ecol. Evol. 2016, 6, 3–22. [Google Scholar] [CrossRef]
- Ciucani, M.M.; Ramos-Madrigal, J.; Hernández-Alonso, G.; Carmagnini, A.; Aninta, S.G.; Scharff-Olsen, C.H.; Lanigan, L.T.; Fracasso, I.; Clausen, C.G.; Aspi, J.; et al. Genomes of the extinct Sicilian wolf reveal a complex history of isolation and admixture with ancient dogs. bioRxiv 2022. Submitted. [Google Scholar] [CrossRef]
- Pierpaoli, M.; Riga, F.; Trocchi, V.; Randi, E. Species distinction and evolutionary relationships of the Italian hare (Lepus corsicanus) as described by mitochondrial DNA sequencing. Mol. Ecol. 1999, 8, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Trucchi, E.; Sbordoni, V. Unveiling an ancient biological invasion: Molecular analysis of an old European alien, the crested porcupine (Hystrix cristata). BMC Evol. Biol. 2009, 9, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Valvo, M.; Russo, R.; Mancuso, F.P.; Palla, F. Mtdna diversity in a rabbit population from Sicily (Italy). Turk. J. Zool. 2017, 41, 645–653. [Google Scholar] [CrossRef]
- Sato, T.; Abramov, A.V.; Raichev, E.G.; Kosintsev, P.A.; Väinölä, R.; Murakami, T.; Kaneko, Y.; Masuda, R. Phylogeography and population history of the least weasel (Mustela nivalis) in the Palearctic based on multilocus analysis. J. Zool. Syst. Evol. Res. 2019, 58, 408–426. [Google Scholar] [CrossRef]
- Frati, F.; Hartl, G.B.; Lovari, S.; Delibes, M.; Markov, G. Quaternary radiation and genetic structure of the red fox Vulpes vulpes in the Mediterranean Basin, as revealed by allozymes and mitochondrial DNA. J. Zool. 1998, 245, 43–51. [Google Scholar] [CrossRef]
- Randi, E. Phylogeography of South European mammals. In Phylogeography of Southern European Refugia; Weiss, S., Ferrand, N., Eds.; Springer: New York, NY, USA, 2007; pp. 101–126. [Google Scholar]
- Schmitt, T.; Fritz, U.; Delfino, M.; Ulrich, W.; Habel, J.C. Biogeography of Italy revisited: Genetic lineages confirm major phylogeographic patterns and a pre-Pleistocene origin of its biota. Front. Zool. 2021, 18, 34. [Google Scholar] [CrossRef]
- Tagliacozzo, A. Archeozoologia della Grotta dell’Uzzo, Sicilia. Suppl. Boll. Soc. Paleontol. Ital. 1993, 84, 1–278. [Google Scholar]
- Petruso, D.; Sara’, M.; Surdi, G.; Masini, F. Le faune a mammiferi della Sicilia tra il Tardiglaciale e l’Olocene. Biogeogr. J. Integr. Biogeogr. 2011, 30, 27–39. [Google Scholar] [CrossRef]
- D’Amico, M.; Bastianelli, G.; Faraone, P.F.; Lo Valvo, M. The spreading of the invasive Italian wall lizard on Vulcano, the last island inhabited by the critically endangered Aeolian wall lizard. Herpetol. Conserv. Biol. 2018, 13, 146–157. [Google Scholar]
- Vecchioni, L.; Marrone, F.; Arculeo, M.; Fritz, U.; Vamberger, M. Stand out from the crowd: Small-scale genetic structuring in the endemic Sicilian pond turtle. Diversity 2020, 12, 343. [Google Scholar] [CrossRef]
- Barbanera, F.; Zuffi, M.A.L.; Guerrini, M.; Gentilli, A.; Tofanelli, S.; Fasola, M.; Dini, F. Molecular phylogeography of the asp viper Vipera aspis (Linnaeus, 1758) in Italy: Evidence for introgressive hybridization and mitochondrial DNA capture. Mol. Phylogenet. Evol. 2009, 52, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Marzahn, E.; Mayer, W.; Joger, U.; Ilgaz, Ç.; Jablonski, D.; Kindler, C.; Kumlutaş, Y.; Nistri, A.; Schneeweiss, N.; Vamberger, M.; et al. Phylogeography of the Lacerta viridis complex: Mitochondrial and nuclear markers provide taxonomic insights. J. Zool. Syst. Evol. Res. 2016, 54, 85–105. [Google Scholar] [CrossRef]
- Senczuk, G.; Colangelo, P.; De Simone, E.; Aloise, G.; Castiglia, R. A combination of long term fragmentation and glacial persistence drove the evolutionary history of the Italian wall lizard Podarcis siculus. BMC Evol. Biol. 2017, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Schultze, N.; Spitzweg, C.; Corti, C.; Delaugerre, M.; Di Nicola, M.R.; Geniez, P.; Lapini, L.; Liuzzi, C.; Lunghi, E.; Novarini, N.; et al. Mitochondrial ghost lineages blur phylogeography and taxonomy of Natrix helvetica and N. natrix in Italy and Corsica. Zool. Scr. 2020, 49, 395–411. [Google Scholar] [CrossRef] [Green Version]
- Chiocchio, A.; Arntzen, J.W.; Martínez-Solano, I.; de Vries, W.; Bisconti, R.; Pezzarossa, A.; Maiorano, L.; Canestrelli, D. Reconstructing hotspots of genetic diversity from glacial refugia and subsequent dispersal in Italian common toads (Bufo bufo). Sci. Rep. 2021, 11, 260. [Google Scholar] [CrossRef]

| Sampling Date | Location | Province | Coordinates | Elevation (m a.s.l.) | “mtDNA I” A.N. | “mtDNA II” A.N. |
|---|---|---|---|---|---|---|
| 25 August 2016 | SS186, Monreale | Palermo | 38.076972° N, 13.290619° E | 245 | OP374121 | OP363871 |
| 22 July 2021 | SP157, C.da Calagni | Messina | 38.030690° N, 14.821550° E | 494 | OP374122 | OP363872 |
| 2 August 2021 | SS121, C.da Misericordia | Enna | 37.580474° N, 14.283588° E | 663 | OP374123 | OP363873 |
| 22 May 2022 | SS624, C.da Strasatto | Palermo | 38.008194° N, 13.242387° E | 779 | OP374124 | OP3638714 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vecchioni, L.; Marrone, F.; Costa, S.; Muscarella, C.; Carra, E.; Arizza, V.; Arculeo, M.; Faraone, F.P. The European Pine Marten Martes martes (Linnaeus, 1758) Is Autochthonous in Sicily and Constitutes a Well-Characterised Major Phylogroup within the Species (Carnivora, Mustelidae). Animals 2022, 12, 2546. https://doi.org/10.3390/ani12192546
Vecchioni L, Marrone F, Costa S, Muscarella C, Carra E, Arizza V, Arculeo M, Faraone FP. The European Pine Marten Martes martes (Linnaeus, 1758) Is Autochthonous in Sicily and Constitutes a Well-Characterised Major Phylogroup within the Species (Carnivora, Mustelidae). Animals. 2022; 12(19):2546. https://doi.org/10.3390/ani12192546
Chicago/Turabian StyleVecchioni, Luca, Federico Marrone, Simone Costa, Calogero Muscarella, Elena Carra, Vincenzo Arizza, Marco Arculeo, and Francesco Paolo Faraone. 2022. "The European Pine Marten Martes martes (Linnaeus, 1758) Is Autochthonous in Sicily and Constitutes a Well-Characterised Major Phylogroup within the Species (Carnivora, Mustelidae)" Animals 12, no. 19: 2546. https://doi.org/10.3390/ani12192546
APA StyleVecchioni, L., Marrone, F., Costa, S., Muscarella, C., Carra, E., Arizza, V., Arculeo, M., & Faraone, F. P. (2022). The European Pine Marten Martes martes (Linnaeus, 1758) Is Autochthonous in Sicily and Constitutes a Well-Characterised Major Phylogroup within the Species (Carnivora, Mustelidae). Animals, 12(19), 2546. https://doi.org/10.3390/ani12192546






