Effects of Crotonylation on Reprogramming of Cashmere Goat Somatic Cells with Different Differentiation Degrees
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of MDSCs
2.2. FFC and EFC Cultures
2.3. Cell Immunofluorescence
2.4. Growth Curve of MDSCs, FFCs, and EFCs
2.5. Western Blot
2.6. Cell Viability Assay
2.7. Screening Optimal Incubation Time and NaCr Concentration
2.8. Cell Cycle and Apoptosis Analysis
2.9. Quantitative Real-Time PCR (qRT-PCR)
2.10. Oocyte Collection and IVM
2.11. IVF
2.12. SCNT
2.13. Immunofluorescence Analysis of Embryo
2.14. Statistical Analysis
3. Results
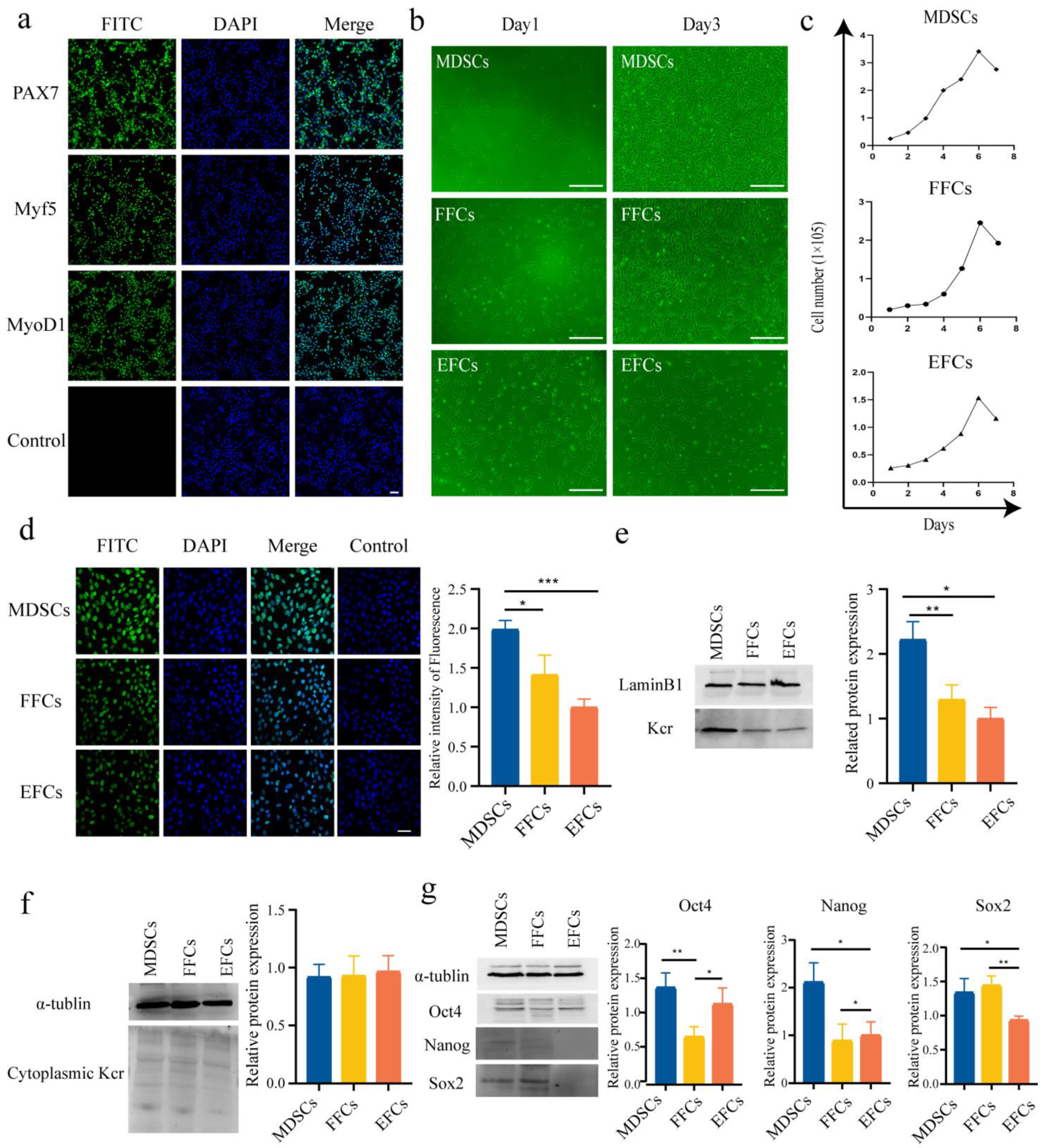
3.1. Somatic Cell Preparation and Culture
3.2. Detection of Kcr Level and of Pluripotency Gene Expression in Somatic Cells
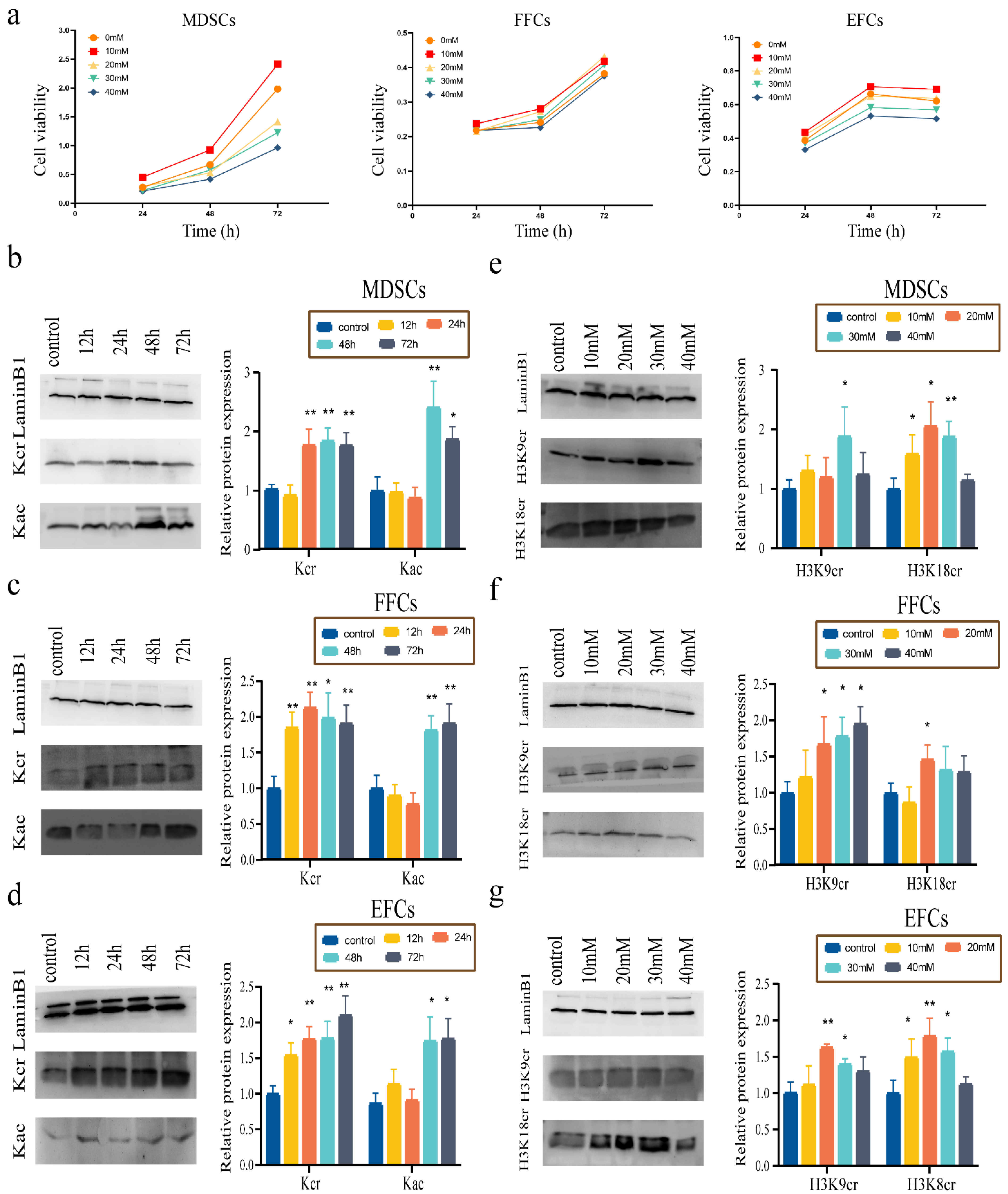
3.3. Effect of NaCr on Cell Viability and Screening of Optimal Incubation Time and Concentration
3.4. Effect of Optimal Incubation Time and NaCr Concentration on Apoptosis and Cell Cycle in Somatic Cells
3.5. Effect of NaCr on the Expression of Key Pluripotency Genes in Somatic Cells
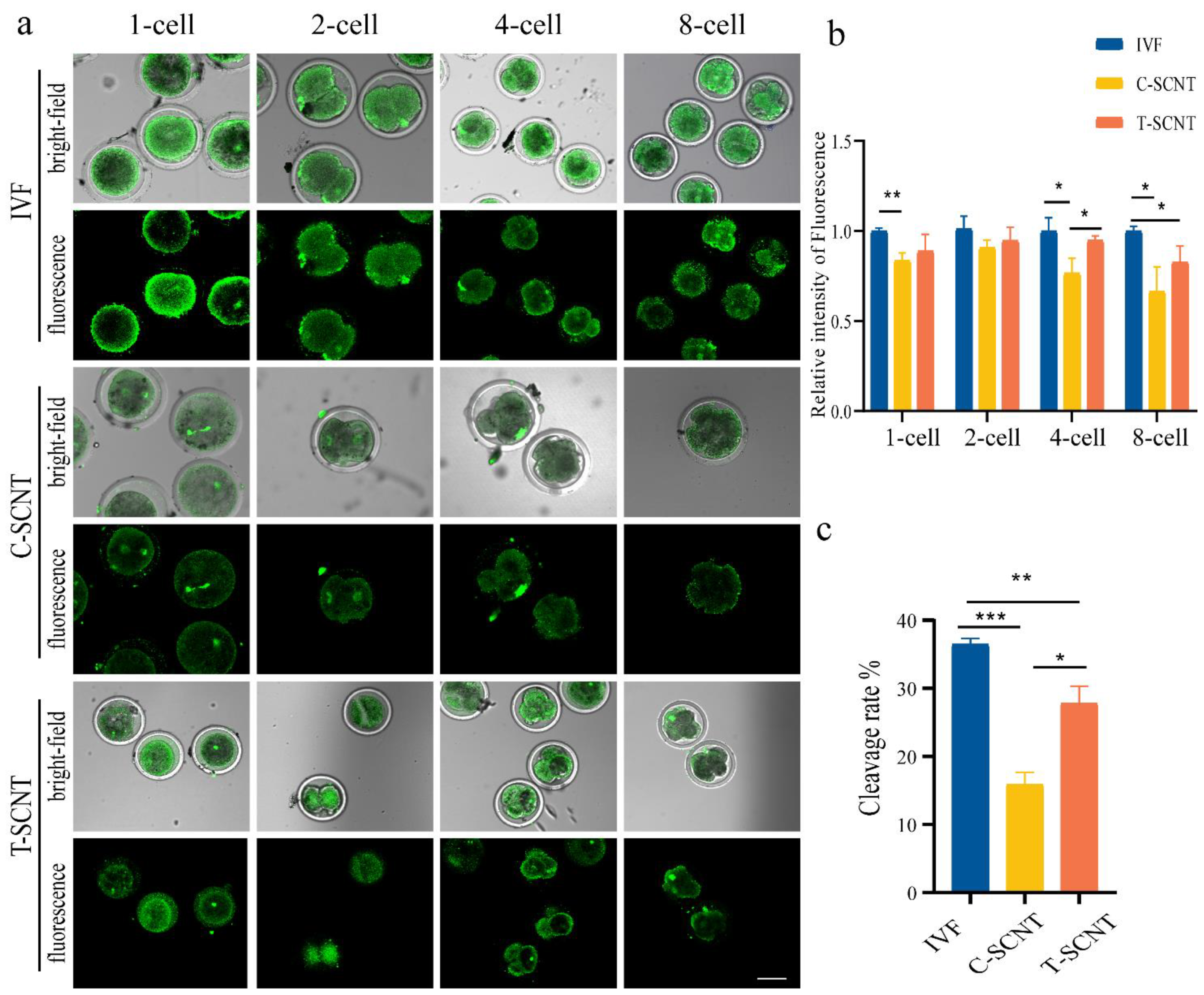
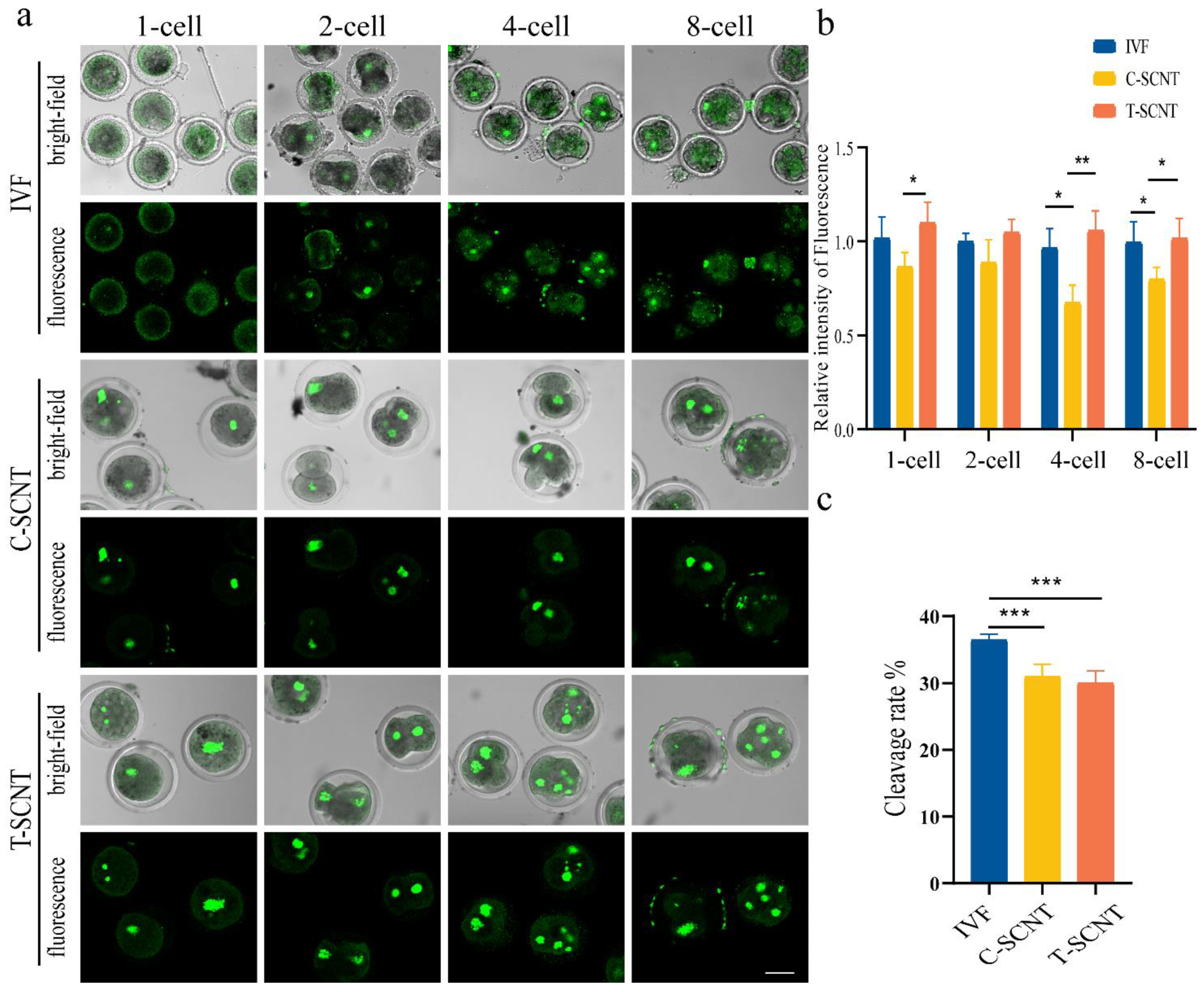
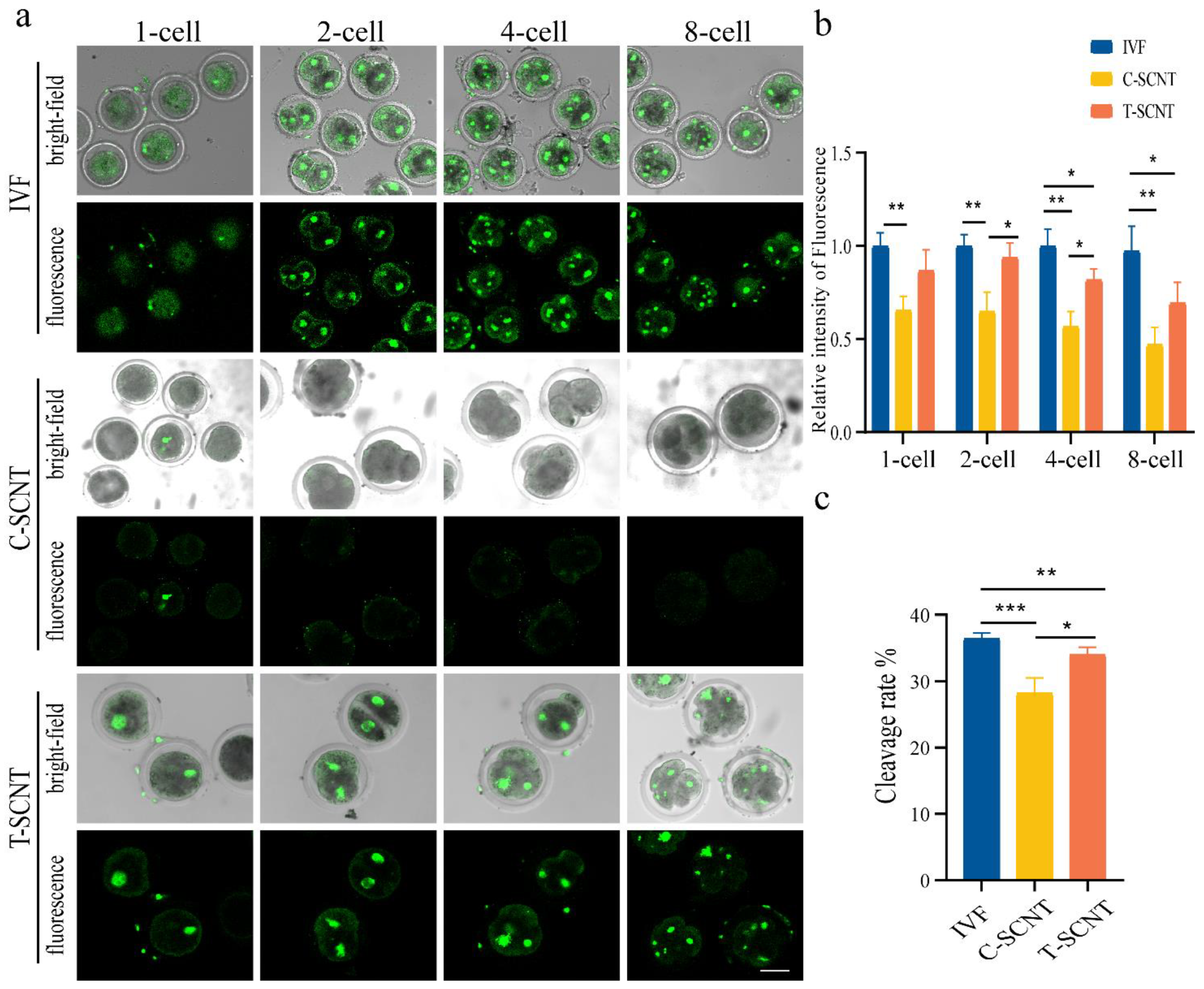
3.6. Comparison of Histone Kcr Modification Levels between IVF Embryos and Cloned Embryos
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Ogura, A.; Inoue, K.; Wakayama, T. Recent advancements in cloning by somatic cell nuclear transfer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20110329. [Google Scholar] [CrossRef] [Green Version]
- Shi, W.; Zakhartchenko, V.; Wolf, E. Epigenetic reprogramming in mammalian nuclear transfer. Differentiation 2003, 71, 91–113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Whyte, J.; Prather, R.S. Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 2010, 341, 13–21. [Google Scholar] [CrossRef]
- Deng, M.; Liu, Z.; Chen, B.; Wan, Y.; Yang, H.; Zhang, Y.; Cai, Y.; Zhou, J.; Wang, F. Aberrant DNA and histone methylation during zygotic genome activation in goat cloned embryos. Theriogenology 2020, 148, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Liu, Z.; Ren, C.; Zhang, G.; Pang, J.; Zhang, Y.; Wang, F.; Wan, Y. Long noncoding RNAs exchange during zygotic genome activation in goat. Biol. Reprod. 2018, 99, 707–717. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Tang, X.C.; Xie, W.H.; Zhou, Y.; Li, D.; Yao, C.G.; Zhou, Y.; Zhu, J.G.; Lai, L.X.; Ouyang, H.S.; et al. Histone Deacetylase Inhibitor Significantly Improved the Cloning Efficiency of Porcine Somatic Cell Nuclear Transfer Embryos. Cell. Reprogram. 2011, 13, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.H.; Li, Z.C.; Yu, B.; He, X.Y.; Shi, J.S.; Zhou, R.; Liu, D.W.; Wu, Z.F. Effects of DNMT1 and HDAC Inhibitors on Gene-Specific Methylation Reprogramming during Porcine Somatic Cell Nuclear Transfer. PLoS ONE 2013, 8, e64705. [Google Scholar] [CrossRef] [Green Version]
- Kishigami, S.; Mizutani, E.; Ohta, H.; Hikichi, T.; Thuan, N.V.; Wakayama, S.; Bui, H.T.; Wakayama, T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem. Biophys. Res. Commun. 2006, 340, 183–189. [Google Scholar] [CrossRef]
- Rybouchkin, A.; Kato, Y.; Tsunodaz, Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol. Reprod. 2006, 74, 1083–1089. [Google Scholar] [CrossRef] [Green Version]
- Whitworth, K.M.; Zhao, J.G.; Spate, L.D.; Li, R.F.; Prather, R.S. Scriptaid Corrects Gene Expression of a Few Aberrantly Reprogrammed Transcripts in Nuclear Transfer Pig Blastocyst Stage Embryos. Cell. Reprogram. 2011, 13, 191–204. [Google Scholar] [CrossRef]
- Mao, J.D.; Zhao, M.T.; Whitworth, K.M.; Spate, L.D.; Walters, E.M.; O’Gorman, C.; Lee, K.; Samuel, M.S.; Murphy, C.N.; Wells, K.; et al. Oxamflatin Treatment Enhances Cloned Porcine Embryo Development and Nuclear Reprogramming. Cell. Reprogram. 2015, 17, 28–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, F.L.; Stamms, K.; Bennewitz, R.; Green, A.; Oback, F.; Turner, P.; Wei, J.W.; Oback, B. Targeted histone demethylation improves somatic cell reprogramming into cloned blastocysts but not postimplantation bovine concepti. Biol. Reprod. 2020, 103, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.M.; Wang, Q.Q.; Liu, K.L.; Gao, E.E.; Guan, H.; Hou, J. Treatment of donor cells with recombinant KDM4D protein improves preimplantation development of cloned ovine embryos. Cytotechnology 2018, 70, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.L.; Liu, L.L.; Ye, X.Y.; Fu, H.F.; Sheng, X.Y.; Wang, L.L.; Wang, H.S.; Heng, D.; Liu, L. Functional Oocytes Derived from Granulosa Cells. Cell Rep. 2019, 29, 4256. [Google Scholar] [CrossRef] [Green Version]
- Chuang, C.Y.; Huang, M.C.; Chen, H.F.; Tseng, L.H.; Yu, C.Y.; Stone, L.; Huang, H.P.; Ho, H.N.; Kuo, H.C. Granulosa cell-derived induced pluripotent stem cells exhibit pro-trophoblastic differentiation potential. Stem Cell Res. Ther. 2015, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Montellier, E.; Rousseaux, S.; Zhao, Y.; Khochbin, S. Histone crotonylation specifically marks the haploid male germ cell gene expression program: Post-meiotic male-specific gene expression. Bioessays 2012, 34, 187–193. [Google Scholar] [CrossRef]
- Keskintepe, L.; Darwish, G.M.; Younis, A.I.; Brackett, B.G. In vitro development of morulae from immature caprine oocytes. Zygote 1994, 2, 97–102. [Google Scholar] [CrossRef]
- Li, Z.C.; He, X.Y.; Chen, L.W.; Shi, J.S.; Zhou, R.; Xu, W.H.; Liu, D.W.; Wu, Z.F. Bone Marrow Mesenchymal Stem Cells are an Attractive Donor Cell Type for Production of Cloned Pigs as Well as Genetically Modified Cloned Pigs by Somatic Cell Nuclear Transfer. Cell. Reprogram. 2013, 15, 459–470. [Google Scholar] [CrossRef] [Green Version]
- Gong, G.; Dai, Y.; Zhu, H.; Wang, H.; Wang, L.; Li, R.; Wan, R.; Liu, Y.; Li, N. Generation of cloned calves from different types of somatic cells. Sci. China Ser. C Life Sci. 2004, 47, 470–476. [Google Scholar] [CrossRef]
- Kishi, M.; Itagaki, Y.; Takakura, R.; Imamura, M.; Sudo, T.; Yoshinari, M.; Tanimoto, M.; Yasue, H.; Kashima, N. Nuclear transfer in cattle using colostrum-derived mammary gland epithelial cells and ear-derived fibroblast cells. Theriogenology 2000, 54, 675–684. [Google Scholar] [CrossRef]
- Zhang, W.B.; Ross, P.J.; Ellis, J.; Salter, M.W. Targeting NMDA receptors in neuropsychiatric disorders by drug screening on human neurons derived from pluripotent stem cells. Transl. Psychiatry 2022, 12, 243. [Google Scholar] [CrossRef] [PubMed]
- Blelloch, R.; Wang, Z.; Meissner, A.; Pollard, S.; Smith, A.; Jaenisch, R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells 2006, 24, 2007–2013. [Google Scholar] [CrossRef] [Green Version]
- Lei, L.; Liu, Z.H.; Wang, H.; Kou, Z.H.; Wu, Y.Q.; Xu, Y.; Cheng, Y.; Zhu, Z.Y.; Xia, G.L.; Chen, D.Y. The effects of different donor cells and passages on development of reconstructed embryos. Acta Genet. Sin. 2003, 30, 215–220. [Google Scholar] [PubMed]
- Young, R.A. Control of the embryonic stem cell state. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Oron, E.; Nelson, B.; Razis, S.; Ivanova, N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 2012, 10, 440–454. [Google Scholar] [CrossRef] [Green Version]
- Pan, C.; Jia, W.; Lu, B.; Bishop, C.E. Expression of TAT recombinant Oct4, Sox2, Lin28, and Nanog proteins from baculovirus-infected Sf9 insect cells. Gene 2015, 556, 245–248. [Google Scholar] [CrossRef]
- Grubelnik, G.; Boštjančič, E.; Pavlič, A.; Kos, M.; Zidar, N. NANOG expression in human development and cancerogenesis. Exp. Biol. Med. 2020, 245, 456–464. [Google Scholar] [CrossRef] [Green Version]
- Matoba, S.; Liu, Y.; Lu, F.; Iwabuchi, K.A.; Shen, L.; Inoue, A.; Zhang, Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 2014, 159, 884–895. [Google Scholar] [CrossRef] [Green Version]
- Yamanaka, K.; Sugimura, S.; Wakai, T.; Kawahara, M.; Sato, E. Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J. Reprod. Dev. 2009, 55, 638–644. [Google Scholar] [CrossRef] [Green Version]
- Kamimura, S.; Inoue, K.; Mizutani, E.; Kim, J.M.; Inoue, H.; Ogonuki, N.; Miyamoto, K.; Ihashi, S.; Itami, N.; Wakayama, T.; et al. Improved development of mouse somatic cell nuclear transfer embryos by chlamydocin analogues, class I and IIa histone deacetylase inhibitors. Biol. Reprod. 2021, 105, 543–553. [Google Scholar] [CrossRef]
- Sabari, B.R.; Tang, Z.Y.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.Z.; Shimada, M.; Cross, J.R.; Zhao, Y.M.; et al. Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol. Cell. 2015, 58, 203–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, M.J.; Luo, H.; Lee, S.; Jin, F.L.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.Y.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 Histone Marks and Histone Lysine Crotonylation as a New Type of Histone Modification. Cell 2011, 146, 1015–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crespo, M.; Damont, A.; Blanco, M.; Lastrucci, E.; El Kennani, S.; Ialy-Radio, C.; El Khattabi, L.; Terrier, S.; Louwagie, M.; Kieffer-Jaquinod, S.; et al. Multi-omic analysis of gametogenesis reveals a novel signature at the promoters and distal enhancers of active genes. Nucleic Acids Res. 2020, 48, 4115–4138. [Google Scholar] [CrossRef]
- Wei, W.; Mao, A.; Tang, B.; Zeng, Q.; Gao, S.; Liu, X.; Lu, L.; Li, W.; Du, J.X.; Li, J.; et al. Large-Scale Identification of Protein Crotonylation Reveals Its Role in Multiple Cellular Functions. J. Proteome Res. 2017, 16, 1743–1752. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Liu, X.; Chen, J.; Gao, S.; Lu, L.; Zhang, H.; Ding, G.; Wang, Z.; Chen, Z.; Shi, T.; et al. Class I histone deacetylases are major histone decrotonylases: Evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017, 27, 898–915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, Y.; Hirayama, M.; Takeda, K.; Tukamoto, N.; Sakata, O.; Kaeriyama, H.; Geshi, M. Effect of synchronization of donor cells in early G1-phase using shake-off method on developmental potential of somatic cell nuclear transfer embryos in cattle. Anim. Sci. J. 2013, 84, 592–599. [Google Scholar] [CrossRef]
- Jia, J.; Zheng, X.; Hu, G.; Cui, K.; Zhang, J.; Zhang, A.; Jiang, H.; Lu, B.; Yates, J., 3rd; Liu, C.; et al. Regulation of pluripotency and self- renewal of ESCs through epigenetic-threshold modulation and mRNA pruning. Cell 2012, 151, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Saini, M.; Selokar, N.L.; Agrawal, H.; Singla, S.K.; Chauhan, M.S.; Manik, R.S.; Palta, P. Treatment of buffalo (Bubalus bubalis) donor cells with trichostatin A and 5-aza-2′-deoxycytidine alters their growth characteristics, gene expression and epigenetic status and improves the in vitro developmental competence, quality and epigenetic status of cloned embryos. Reprod. Fertil. Dev. 2016, 28, 824–837. [Google Scholar] [CrossRef]
- Jin, L.; Zhu, H.Y.; Guo, Q.; Li, X.C.; Zhang, Y.C.; Cui, C.D.; Li, W.X.; Cui, Z.Y.; Yin, X.J.; Kang, J.D. Effect of histone acetylation modification with MGCD0103, a histone deacetylase inhibitor, on nuclear reprogramming and the developmental competence of porcine somatic cell nuclear transfer embryos. Theriogenology 2017, 87, 298–305. [Google Scholar] [CrossRef]
- Mao, T.C.; Han, C.Q.; Deng, R.Z.; Wei, B.; Meng, P.; Luo, Y.; Zhang, Y. Treating donor cells with 2-PCPA corrects aberrant histone H3K4 dimethylation and improves cloned goat embryo development. Syst. Biol. Reprod. Med. 2018, 64, 174–182. [Google Scholar] [CrossRef]
- Xu, Y.X.; Shi, Z.Y.; Bao, L. An Expanding Repertoire of Protein Acylations. Mol. Cell Proteom. 2022, 21, 100193. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, A.M.; Hylan, D.A.; Ballard, C.B.; Purpera, M.N.; Vaught, T.D.; Lynn, J.W.; Godke, R.A.; Bondioli, K.R. Effect of epigenetic modifications of donor somatic cells on the subsequent chromatin remodeling of cloned bovine embryos. Biol. Reprod. 2008, 78, 832–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, I.H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Black, J.B.; Adler, A.F.; Wang, H.G.; D’Ippolito, A.M.; Hutchinson, H.A.; Reddy, T.E.; Pitt, G.S.; Leong, K.W.; Gersbach, C.A. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell 2016, 19, 406–414. [Google Scholar] [CrossRef] [Green Version]
- Hosseini, S.M.; Dufort, I.; Nieminen, J.; Moulavi, F.; Ghanaei, H.R.; Hajian, M.; Jafarpour, F.; Forouzanfar, M.; Gourbai, H.; Shahverdi, A.H.; et al. Epigenetic modification with trichostatin A does not correct specific errors of somatic cell nuclear transfer at the transcriptomic level; highlighting the non-random nature of oocyte-mediated reprogramming errors. BMC Genom. 2016, 17, 16. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Diaz, M.A.; Che, L.; Albornoz, M.; Seneda, M.M.; Collis, D.; Coutinho, A.R.; El-Beirouthi, N.; Laurin, D.; Zhao, X.; Bordignon, V. Pre- and postimplantation development of swine-cloned embryos derived from fibroblasts and bone marrow cells after inhibition of histone deacetylases. Cell. Reprogram. 2010, 12, 85–94. [Google Scholar] [CrossRef]


| Gene Name | Primer Sequence (5′-3′) | Tm (°C) |
|---|---|---|
| GAPDH | F: TTGTGATGGGCGTGAACC | 50 |
| R: CCCTCCACGATGCCAAA | 48 | |
| Nanog | F: GTCTCTCCTCTTCCTTCCTCCA | 59 |
| R: TCTTCCTTCTCTGTGCTCTCCTC | 57 | |
| Oct4 | F: GCCAAGCTCCTAAAGCAGAAGA | 62 |
| R: AAAGCCTCAAAACGGCAGATAG | 63 | |
| Sox2 | F: CATGATGGAGACGGAACTGG | 55 |
| R: CGGGCTGTTCTTCTGGTTG | 53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Yan, W.; Hao, F.; Hao, L.; Liu, D. Effects of Crotonylation on Reprogramming of Cashmere Goat Somatic Cells with Different Differentiation Degrees. Animals 2022, 12, 2848. https://doi.org/10.3390/ani12202848
Li W, Yan W, Hao F, Hao L, Liu D. Effects of Crotonylation on Reprogramming of Cashmere Goat Somatic Cells with Different Differentiation Degrees. Animals. 2022; 12(20):2848. https://doi.org/10.3390/ani12202848
Chicago/Turabian StyleLi, Wennan, Wei Yan, Fei Hao, Lingyun Hao, and Dongjun Liu. 2022. "Effects of Crotonylation on Reprogramming of Cashmere Goat Somatic Cells with Different Differentiation Degrees" Animals 12, no. 20: 2848. https://doi.org/10.3390/ani12202848
APA StyleLi, W., Yan, W., Hao, F., Hao, L., & Liu, D. (2022). Effects of Crotonylation on Reprogramming of Cashmere Goat Somatic Cells with Different Differentiation Degrees. Animals, 12(20), 2848. https://doi.org/10.3390/ani12202848




