Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Culture of Goose Myoblasts
2.2. RNA Extraction, cDNA Synthesis, Fluorescence Quantitative PCR
2.3. Library Construction and Illumine Sequencing
2.4. Bioinformatics Analysis
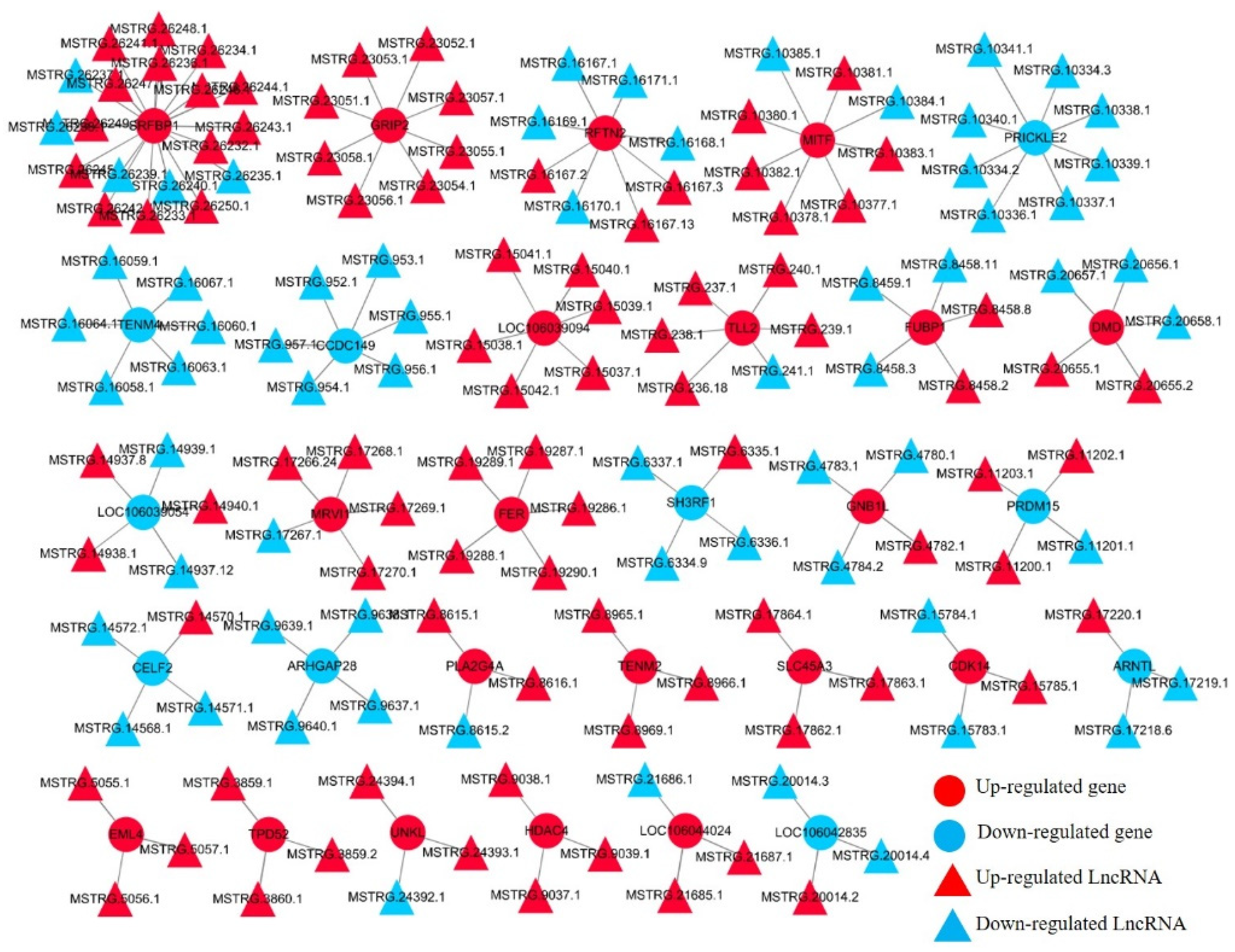
2.5. Construction of DE-lncRNAs-DEGs Interaction Network
2.6. Statistical Analysis of qPCR Results
3. Results
3.1. Isolation and Transcriptome Sequencing of Shitou Goose Myoblasts
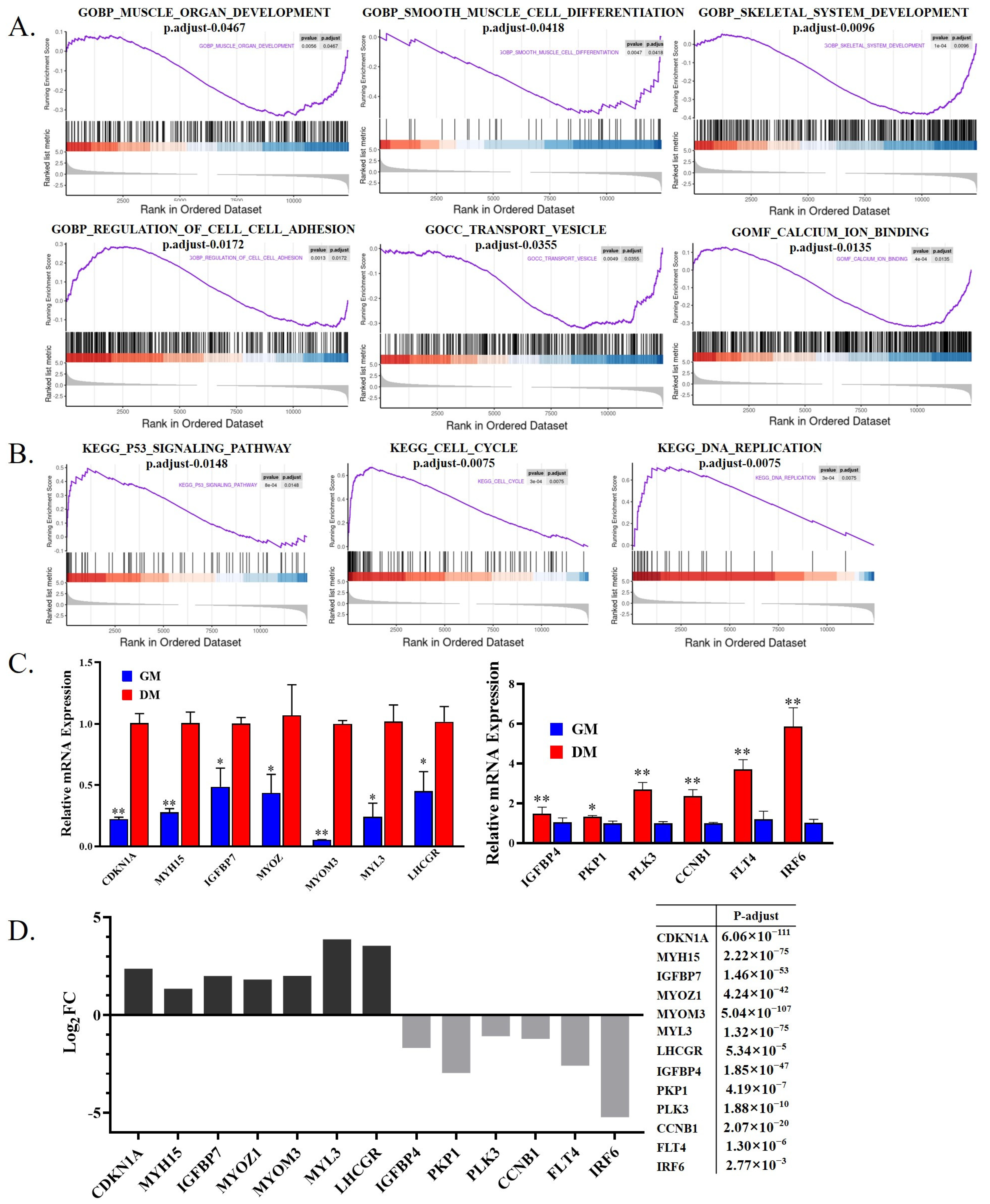
3.2. Functional Analysis of DEGs between Shitou Goose Myoblasts and Myotubes
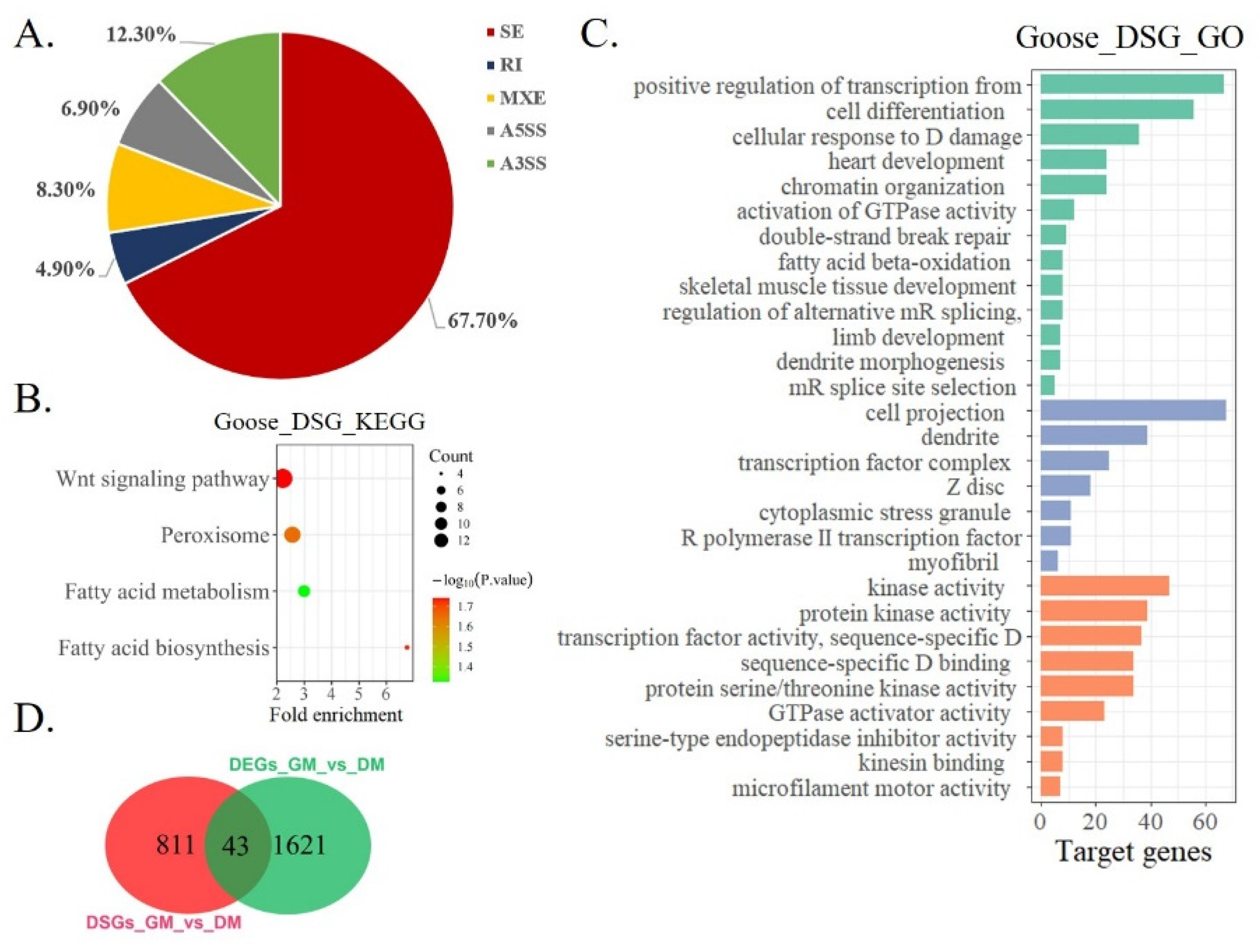
3.3. Differential Splicing Analysis of mRNA during Goose Myoblast Differentiation
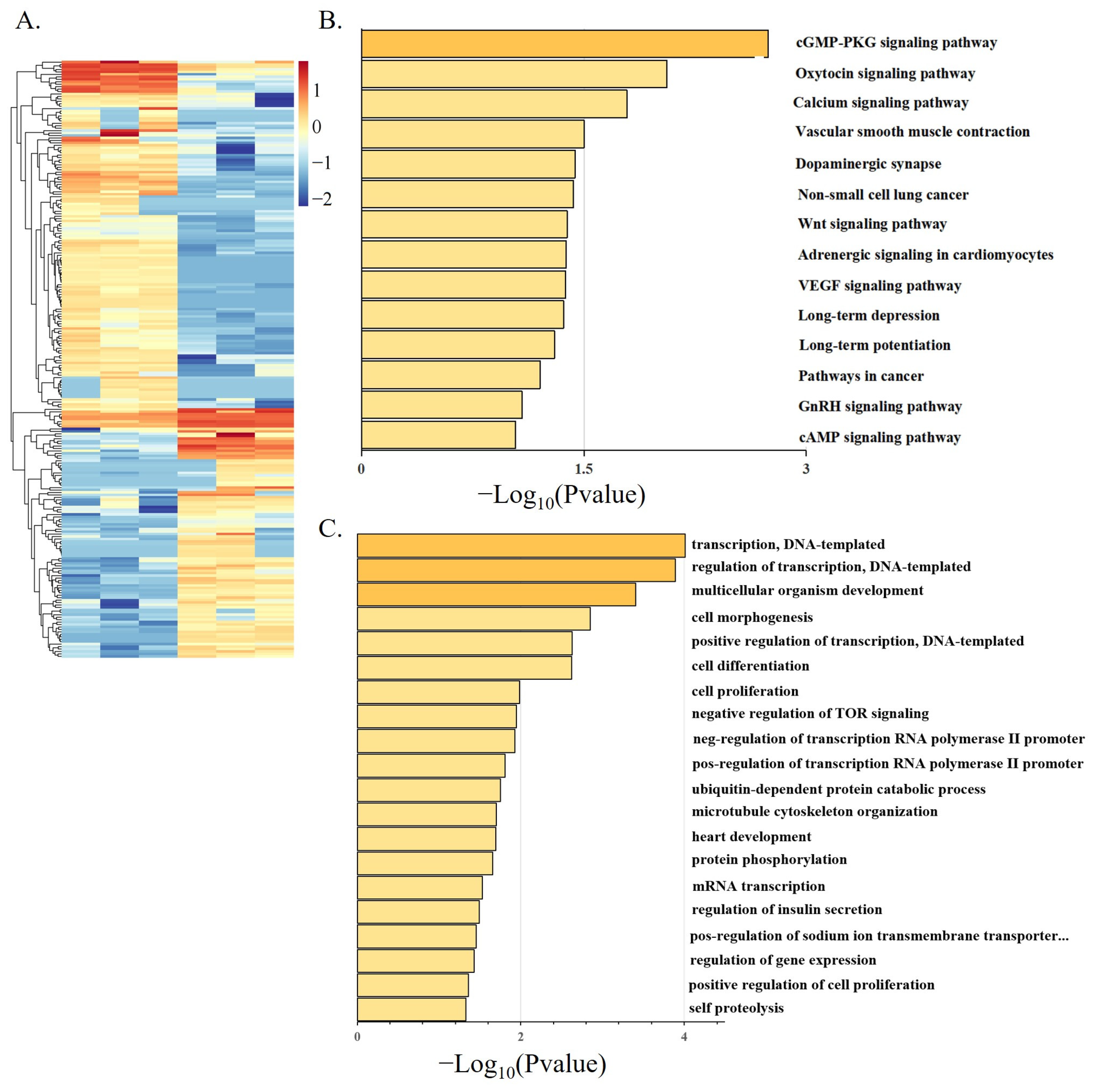
3.4. Differential Expression Analysis of LncRNA during Goose Myoblasts Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henchion, M.; Moloney, A.; Hyland, J.; Zimmermann, J.; McCarthy, S. Review: Trends for meat, milk and egg consumption for the next decades and the role played by livestock systems in the global production of proteins. Animal 2021, 15, 100287. [Google Scholar] [CrossRef]
- Luo, W.; Lin, Z.; Chen, J.; Chen, G.; Zhang, S.; Liu, M.; Li, H.; He, D.; Liang, S.; Luo, Q.; et al. TMEM182 interacts with integrin beta 1 and regulates myoblast differentiation and muscle regeneration. J. Cachex. Sarcopenia Muscle 2021, 12, 1704–1723. [Google Scholar] [CrossRef]
- Braun, T.; Gautel, M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol. 2011, 12, 349–361. [Google Scholar] [CrossRef]
- Wang, S.; Jin, J.; Xu, Z.; Zuo, B. Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production. Cells 2019, 8, 1107. [Google Scholar] [CrossRef]
- Ye, C.; Zhong, R. Study on the growth model of lion headed geese. J. Shihezi Univ. (Nat. Sci. Ed.) 2005, 7, 694–697. [Google Scholar]
- Hou, S.; Liu, L. Present situation, future development trend and suggestions of waterfowl industry in 2020. Chin. J. Anim. Husb. 2021, 57, 235–239. [Google Scholar]
- Tang, J.; Guo, M.; Fu, J.; Ouyang, H.; Tian, Y.; Shen, X.; Yunmao, H. Polymorphism analysis and expression patterns of the IGF1 gene in the Shitou goose. Arch. Anim. Breed. 2021, 64, 315–323. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Li, X.; Shen, X.; Xu, D.; Tian, Y.; Huang, Y. Transcriptomic investigation of embryonic pectoral muscle reveals increased myogenic processes in Shitou geese compared to Wuzong geese. Br. Poult. Sci. 2021, 62, 650–657. [Google Scholar] [CrossRef]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Ben-Dov, C.; Hartmann, B.; Lundgren, J.; Valcarcel, J. Genome-wide analysis of alternative pre-mRNA splicing. J. Biol. Chem. 2008, 283, 1229–1233. [Google Scholar] [CrossRef]
- Matlin, A.J.; Clark, F.; Smith, C.W.J. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005, 6, 386–398. [Google Scholar] [CrossRef]
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003, 72, 291–336. [Google Scholar] [CrossRef]
- Johnson, J.M.; Castle, J.; Garrett-Engele, P.; Kan, Z.; Loerch, P.M.; Armour, C.D.; Santos, R.; Schadt, E.E.; Stoughton, R.; Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 2003, 302, 2141–2144. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef]
- Dey, B.K.; Pfeifer, K.; Dutta, A. TheH19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Gene Dev. 2014, 28, 491–501. [Google Scholar] [CrossRef]
- Zheng, L.; Liu, X.; Chen, P.; Xiao, W. [Corrigendum] Expression and role of lncRNAs in the regeneration of skeletal muscle following contusion injury. Exp. Ther. Med. 2019, 19, 797. [Google Scholar] [CrossRef]
- Gong, C.; Li, Z.; Ramanujan, K.; Clay, I.; Zhang, Y.; Lemire-Brachat, S.; Glass, D.J. A Long Non-coding RNA, LncMyoD, Regulates Skeletal Muscle Differentiation by Blocking IMP2-Mediated mRNA Translation. Dev. Cell 2015, 34, 181–191. [Google Scholar] [CrossRef]
- Ju, X.; Liu, Y.; Shan, Y.; Ji, G.; Zhang, M.; Tu, Y.; Zou, J.; Chen, X.; Geng, Z.; Shu, J. Analysis of potential regulatory LncRNAs and CircRNAs in the oxidative myofiber and glycolytic myofiber of chickens. Sci. Rep. 2021, 11, 20861. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, T.; Wu, C.; Wang, S.; Wang, Y.; Li, H.; Wang, N.; Ouellette, M.M. Immortalization of chicken preadipocytes by retroviral transduction of chicken TERT and TR. PLoS ONE 2017, 12, e177348. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Kong, L.; Zhang, Y.; Ye, Z.-Q.; Liu, X.-Q.; Zhao, S.-Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef]
- Sun, L.; Luo, H.; Bu, D.; Zhao, G.; Yu, K.; Zhang, C.; Liu, Y.; Chen, R.; Zhao, Y. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.-X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Navarro Gonzalez, J.; Zweig, A.S.; Speir, M.L.; Schmelter, D.; Rosenbloom, K.R.; Raney, B.J.; Powell, C.C.; Nassar, L.R.; Maulding, N.D.; Lee, C.M.; et al. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021, 49, D1046–D1057. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Raguraman, P.; Balachandran, A.A.; Chen, S.; Diermeier, S.D.; Veedu, R.N. Antisense Oligonucleotide-Mediated Splice Switching: Potential Therapeutic Approach for Cancer Mitigation. Cancers 2021, 13, 5555. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Beutler, B.; Zhang, D. Emerging roles of spliceosome in cancer and immunity. Protein Cell. 2021, 13, 559–579. [Google Scholar] [CrossRef]
- Verta, J.P.; Jacobs, A. The role of alternative splicing in adaptation and evolution. Trends Ecol. Evol. 2021, 37, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Schaefke, B.; Li, Y.; Jia, F.; Sun, W.; Li, G.; Liang, W.; Reif, T.; Heyd, F.; Gao, Q.; et al. Mammalian splicing divergence is shaped by drift, buffering in trans, and a scaling law. Life Sci. Alliance 2022, 5, e202101333. [Google Scholar] [CrossRef] [PubMed]
- Archacka, K.; Ciemerych, M.A.; Florkowska, A.; Romanczuk, K. Non-Coding RNAs as Regulators of Myogenesis and Postexercise Muscle Regeneration. Int. J. Mol. Sci. 2021, 22, 11568. [Google Scholar] [CrossRef]
- Liu, Q.; Deng, J.; Qiu, Y.; Gao, J.; Li, J.; Guan, L.; Lee, H.; Zhou, Q.; Xiao, J. Non-coding RNA basis of muscle atrophy. Mol. Ther. Nucleic Acids 2021, 26, 1066–1078. [Google Scholar] [CrossRef]
- Mbadhi, M.N.; Tang, J.-M.; Zhang, J.-X. Histone Lysine Methylation and Long Non-Coding RNA: The New Target Players in Skeletal Muscle Cell Regeneration. Front. Cell. Dev. Biol. 2021, 9, 759237. [Google Scholar] [CrossRef]
- Chen, S.; Chen, K.; Xu, J.; Li, F.; Ding, J.; Ma, Z.; Li, G.; Li, H. Insights Into mRNA and Long Non-coding RNA Profiling RNA Sequencing in Uterus of Chickens with Pink and Blue Eggshell Colors. Front. Veter. Sci. 2021, 8, 736387. [Google Scholar] [CrossRef]
- Suchocki, T.; Czech, B.; Dunislawska, A.; Slawinska, A.; Derebecka, N.; Wesoly, J.; Siwek, M.; Szyda, J. SNP prioritization in targeted sequencing data associated with humoral immune responses in chicken. Poult. Sci. 2021, 100, 101433. [Google Scholar] [CrossRef]
- Guo, G.; Sun, L.; Sun, M.; Xu, H. LncRNA SLC8A1-AS1 protects against myocardial damage through activation of cGMP-PKG signaling pathway by inhibiting SLC8A1 in mice models of myocardial infarction. J. Cell. Physiol. 2019, 234, 9019–9032. [Google Scholar] [CrossRef]
- Zou, L.; Xia, P.; Chen, L.; Hou, Y. XIST knockdown suppresses vascular smooth muscle cell proliferation and induces apoptosis by regulating miR-1264/WNT5A/β-catenin signaling in aneurysm. Biosci. Rep. 2021, 41, BSR20201810. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Chen, J.; Li, L.; Ren, X.; Cheng, T.; Lu, S.; Lawal, R.A.; Nie, Q.; Zhang, X.; Hanotte, O. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fiber hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell. Death Differ. 2019, 26, 426–442. [Google Scholar] [CrossRef] [PubMed]
- Picard, B.; Berri, C.; Lefaucheur, L.; Molette, C.; Sayd, T.; Terlouw, C. Skeletal muscle proteomics in livestock production. Brief. Funct. Genom. 2010, 9, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Bradley, J.S.; Siegel, P.B.; Yang, N.; Johnson, S.E. Selection for divergent body size alters rates of embryonic skeletal muscle formation and muscle gene expression patterns. Dev. Growth Differ. 2015, 57, 614–624. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Huang, Y.; Li, X.; Shen, X.; Fu, X.; Jiang, D.; Ouyang, H.; Fan, D.; Huang, X.; Zhang, X. Culture and identification of goose myoblasts in vitro and the effect of temperature on cell proliferation and differentiation. J. Zhongkai Agric. Eng. Coll. 2020, 33, 21–27. [Google Scholar]
- Owen, K.L.; Brockwell, N.K.; Parker, B.S. JAK-STAT Signaling: A Double-Edged Sword of Immune Regulation and Cancer Progression. Cancers 2019, 11, 2002. [Google Scholar] [CrossRef]
- He, H.; Yin, H.; Yu, X.; Zhang, Y.; Ma, M.; Li, D.; Zhu, Q. PDLIM5 Affects Chicken Skeletal Muscle Satellite Cell Proliferation and Differentiation via the p38-MAPK Pathway. Animals 2021, 11, 1016. [Google Scholar] [CrossRef]
- Yin, H.; Zhao, J.; He, H.; Chen, Y.; Wang, Y.; Li, D.; Zhu, Q. Gga-miR-3525 Targets PDLIM3 through the MAPK Signaling Pathway to Regulate the Proliferation and Differentiation of Skeletal Muscle Satellite Cells. Int. J. Mol. Sci. 2020, 21, 5573. [Google Scholar] [CrossRef]
- Huang, W.; Guo, L.; Zhao, M.; Zhang, D.; Xu, H.; Nie, Q. The Inhibition on MDFIC and PI3K/AKT Pathway Caused by miR-146b-3p Triggers Suppression of Myoblast Proliferation and Differentiation and Promotion of Apoptosis. Cells 2019, 8, 656. [Google Scholar] [CrossRef]
- Huang, H.; Liu, L.; Li, C.; Liang, Z.; Huang, Z.; Wang, Q.; Li, S.; Zhao, Z. Fat mass- and obesity-associated (FTO) gene promoted myoblast differentiation through the focal adhesion pathway in chicken. 3 Biotech 2020, 10, 403. [Google Scholar] [CrossRef]
- Han, S.; Cui, C.; Wang, Y.; He, H.; Liu, Z.; Shen, X.; Chen, Y.; Li, D.; Zhu, Q.; Yin, H. Knockdown of CSRP3 inhibits differentiation of chicken satellite cells by promoting TGF-beta/Smad3 signaling. Gene 2019, 707, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Moresi, V.; Adamo, S.; Berghella, L. The JAK/STAT Pathway in Skeletal Muscle Pathophysiology. Front. Physiol. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Addinsall, A.B.; Cacciani, N.; Akkad, H. JAK/STAT inhibition augments soleus muscle function in a rat model of critical illness myopathy via regulation of complement C3/3R. J. Physiol. 2021, 599, 2869–2886. [Google Scholar] [CrossRef] [PubMed]
- Mathews, E.S.; Appel, B. Cholesterol Biosynthesis Supports Myelin Gene Expression and Axon Ensheathment through Modulation of P13K/Akt/mTor Signaling. J. Neurosci. 2016, 36, 7628–7639. [Google Scholar] [CrossRef]
- Saahene, R.O.; Agbo, E.; Barnes, P. A Review: Mechanism of Phyllanthus urinaria in Cancers—NF-κB, P13K/AKT, and MAPKs Signaling Activation. Evid.-Based Complement. Altern. Med. 2021, 2021, 4514342. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, G.; Yu, K.; Zhang, X.; Jiang, A. MiRNA-615-3p Alleviates Oxidative Stress Injury of Human Cardiomyocytes Via PI3K/Akt Signaling by Targeting MEF2A. Anatol. J. Cardiol. 2022, 26, 373–381. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lonnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Maik-Rachline, G.; Wortzel, I.; Seger, R. Alternative Splicing of MAPKs in the Regulation of Signaling Specificity. Cells 2021, 10, 3466. [Google Scholar] [CrossRef]
- Idris, M.; Harmston, N.; Petretto, E.G.; Madan, B.; Virshup, D.M. Broad regulation of gene isoform expression by Wnt signaling in cancer. RNA 2019, 25, 1696–1713. [Google Scholar] [CrossRef]
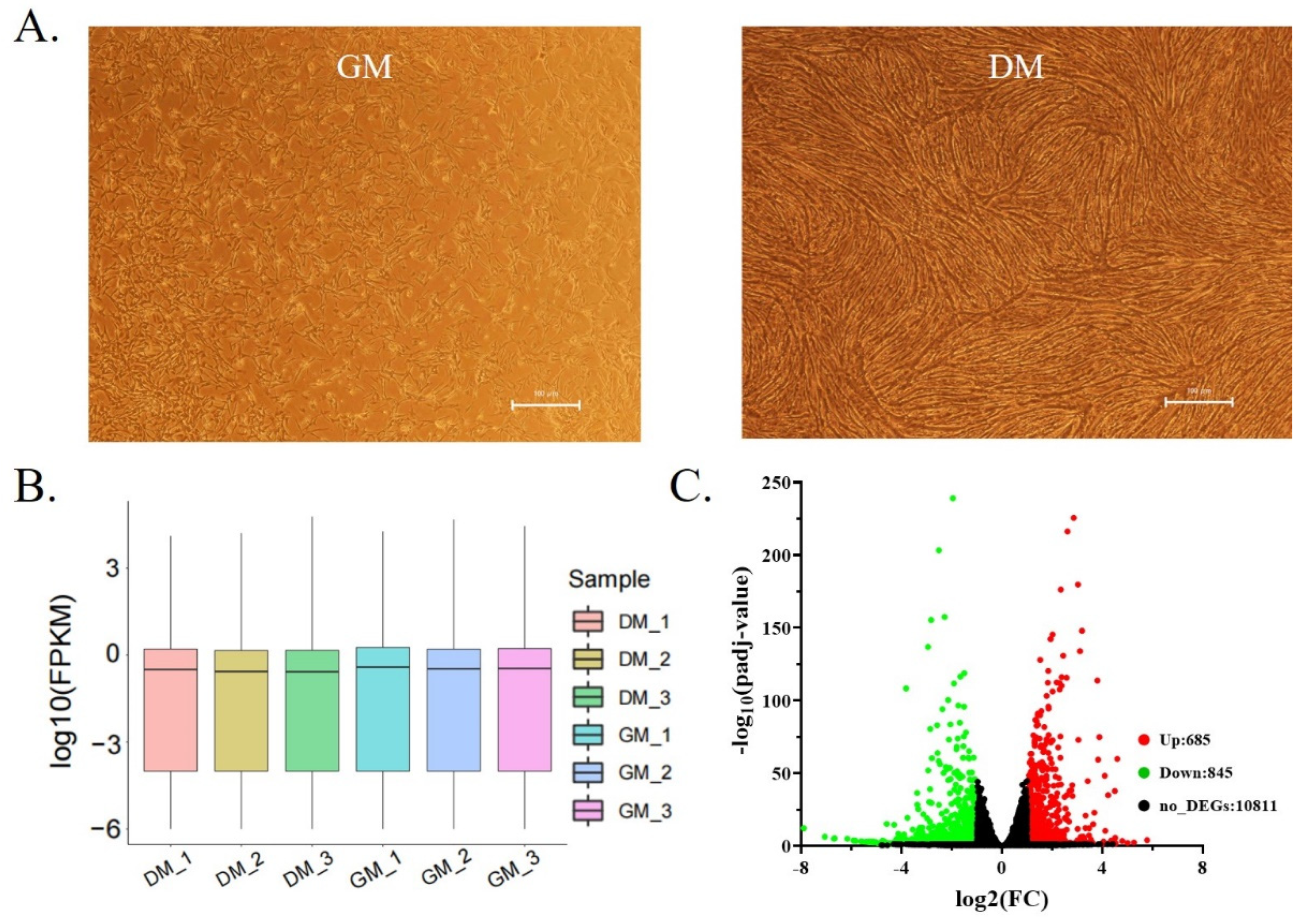

| Samples | Raw Data (Read) | Valid Data (Read) | Mapped Reads | Unique Mapped Reads | Q30% | GC Content % |
|---|---|---|---|---|---|---|
| GM_1 | 70689352 | 63945928 (90.46%) | 60857652 (86.13%) | 57736805 (72.17%) | 98.70% | 47% |
| GM_2 | 79467030 | 73247104 (92.17%) | 69218212 (85.90%) | 59177117 (73.44%) | 98.62% | 48% |
| GM_3 | 74221168 | 68726348 (92.60%) | 60857652 (86.13%) | 52009319 (73.61%) | 98.63% | 48% |
| DM_1 | 86604440 | 80002650 (92.38%) | 55637470 (87.01%) | 47301347 (73.97%) | 98.47% | 47% |
| DM_2 | 89152764 | 80578354 (90.38%) | 63493532 (86.68%) | 53952475 (73.66%) | 98.57% | 47% |
| DM_3 | 78730378 | 70658744 (89.75%) | 59425642 (86.47%) | 50403679 (73.34%) | 98.76% | 47% |
| The Top Ten Genes Up-Regulated during Myoblast Differentiation | The Top Ten Genes Down-Regulated during Myoblast Differentiation | ||||
|---|---|---|---|---|---|
| Name | Log2FC | −log10(padj Value) | Name | Log2FC | −log10(padj Value) |
| NMUR2 | 5.003212 | 2.054442 | ESPN | −5.89484 | 3.649857 |
| SLC14A2 | 4.787548 | 3.415658 | MPZL3 | −5.83747 | 3.542505 |
| FIGF | 4.586098 | 59.866965 | SCARF1 | −5.62258 | 3.159790 |
| MYRFL | 4.519206 | 2.034318 | GFAP | −5.58841 | 3.122162 |
| SHANK1 | 4.505554 | 1.395063 | FAM83A | −5.31 | 2.557658 |
| NYAP2 | 4.505405 | 5.634583 | SFN | −5.28109 | 3.142785 |
| GADL1 | 4.346948 | 3.736134 | IRF6 | −5.22921 | 2.558212 |
| WNT11 | 4.222275 | 35.025755 | TMPRSS4 | −5.14386 | 2.382782 |
| KCNJ16 | 4.113374 | 10.443003 | SDCBP2 | −4.98299 | 2.099845 |
| CKMT2 | 4.087084 | 48.253844 | GJA4 | −4.86464 | 1.923671 |
| Exon Skipping | Intron Retention | Mutually Exclusive Exon | Alternative 5′ Splice Site | Alternative 3′ Splice Site | |||||
|---|---|---|---|---|---|---|---|---|---|
| Name | LncLevel Diff. | Name | LncLevel Diff. | Name | LncLevel Diff. | Name | LncLevel Diff. | Name | LncLevel Diff. |
| SORBS2 | 0.508 | LOC106031985 | 0.508 | SORBS2 | 0.721 | SORBS2 | 0.574 | RBM27 | 0.719 |
| KCTD17 | 0.788 | LOC106047490 | 0.434 | KLF12 | 0.667 | GZF1 | 0.551 | PTPN3 | 0.667 |
| AP1S2 | 0.755 | MOB3C | 0.425 | LOC106042308 | 0.464 | ARPP21 | 0.551 | RAPGEF1 | 0.603 |
| REEP1 | 0.659 | ZNF644 | 0.415 | HENMT1 | 0.436 | SYNCRIP | 0.537 | LOC106032270 | 0.59 |
| CAMSAP2 | 0.627 | SLC39A3 | 0.406 | ACSL6 | 0.431 | LOC106031985 | 0.465 | MPP3 | 0.567 |
| PBX3 | 0.625 | TMEM40 | 0.382 | NCAM1 | 0.423 | LOC106031985 | 0.463 | PRLR | 0.55 |
| FAM76B | 0.624 | IRF1 | 0.376 | ABLIM3 | 0.41 | PLAT | 0.448 | CYSLTR2 | 0.535 |
| NCAM1 | 0.618 | NFKBIZ | 0.343 | FHL5 | 0.365 | SLC39A10 | 0.441 | PRLR | 0.529 |
| TPD52L1 | 0.607 | LOC106044164 | 0.336 | PCNXL2 | 0.36 | LOC106032100 | 0.441 | PRLR | 0.523 |
| EHMT1 | 0.606 | SLC6A1 | 0.333 | EPHB6 | 0.356 | HDAC9 | 0.439 | LOC106031985 | 0.518 |
| GO_Term | DS-Gene | Count | FDR (Adjusted p-Value) |
|---|---|---|---|
| extracellular matrix | SMOC2, POSTN, COL26A1, ADAMTS13, COL24A1, ELN, TNC | 7 | 0.00062 |
| Costamere | SVIL, CMYA5, DMD | 3 | 0.01734 |
| actin cytoskeleton | SVIL, ABLIM1, MYO1C, ABLIM2, DST | 5 | 0.01997 |
| actin binding | SVIL, ABLIM1, MYO1C, ABLIM2, DST, DMD | 6 | 0.03799 |
| extracellular matrix structural constituent | POSTN, COL24A1, ELN, TNC | 4 | 0.04167 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Zhang, S.; Chen, G.; Deng, X.; Zhang, D.; Wen, H.; Yin, Y.; Lin, Z.; Zhang, X.; Luo, W. Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts. Animals 2022, 12, 2956. https://doi.org/10.3390/ani12212956
Chen J, Zhang S, Chen G, Deng X, Zhang D, Wen H, Yin Y, Lin Z, Zhang X, Luo W. Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts. Animals. 2022; 12(21):2956. https://doi.org/10.3390/ani12212956
Chicago/Turabian StyleChen, Jiahui, Shuai Zhang, Genghua Chen, Xianqi Deng, Danlu Zhang, Huaqiang Wen, Yunqian Yin, Zetong Lin, Xiquan Zhang, and Wen Luo. 2022. "Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts" Animals 12, no. 21: 2956. https://doi.org/10.3390/ani12212956
APA StyleChen, J., Zhang, S., Chen, G., Deng, X., Zhang, D., Wen, H., Yin, Y., Lin, Z., Zhang, X., & Luo, W. (2022). Transcriptome Sequencing Reveals Pathways Related to Proliferation and Differentiation of Shitou Goose Myoblasts. Animals, 12(21), 2956. https://doi.org/10.3390/ani12212956





