Simple Summary
The SOX6 (sex determining region Y-box 6) gene belongs to one of the transcription factors in the SRY (sex-determining region Y) family, which affects sex determination, embryonic and nervous system development, bone and various organ formation. In the previous study, the whole-genome sequencing was used to detect multiple genes located in the copy number variation region, including SOX6 gene. In this study, we identified the correlation between the growth traits and CNV of SOX6 in 311 Ashidan yaks. The results showed that SOX6-CNV was significantly correlated with the chest girth of the 6-months old yaks (p < 0.05) and 30-months yaks (p < 0.05), and withers height of 6 months yaks (p < 0.05) and 18-months yaks (p < 0.05), suggesting the SOX6-CNV affect growth traits in yaks, and could be new markers for the selection of yak breeding.
Abstract
Copy number variation (CNV) is a fundamental type of structural variation of the genome affecting the economic traits of livestock. The SOX6 gene (sex-determining region Y-box 6), as a transcription factor, has multiple functions with regard to sex determination, embryonic growth, the nervous system development, as well as bone, and various organ formation. This study employed quantitative real-time fluorescence quota PCR (qPCR) for detecting the SOX6-CNV of the 311 Ashidan yaks and analyzed the correlation of the SOX6-CNV with four phenotypes (including body weight, withers height, body length, and chest girth) of the yaks aged 6, 12, 18, and 30 months using ANOVA and multiple comparisons. Furthermore, the SOX6 gene expression was identified in seven different tissues of the yaks. The experiment results demonstrated the expression of SOX6 in each tissue, and the kidney and muscle tissue were found to have higher relative expression levels. Based on the processing by IBM SPSS software, SOX6-CNV was significantly correlated with the chest girth of the 6-months old yaks (p < 0.05) and 30-months yaks (p < 0.05), and withers height of 6 months yaks (p < 0.05) and 18-months yaks (p < 0.05), as well as the normal type of CNV, was chosen for yak breeding. In conclusion, SOX6 might be prominently involved in promoting growth and development of yaks, suggesting that the SOX6 gene can be used in breeding yaks by molecular marker-assisted selection (MAS). The study also offered some important insights into the references and clues for the genetic breeding of yaks.
1. Introduction
Copy number variation (CNV) is defined as a DNA fragment with a variable copy number size of 1 KB or longer than the reference genome, which is a significant portion of the variant in the genome [1]. CNV mainly shows recombination, deletion, and insertion of multiple loci [2]. CNV is currently considered a polymorphic genetic marker of important economic traits or disease susceptibility phenotypic variation in livestock species [3,4]. Studies over the last couple of decades have shown CNV to be compactly related to important growth traits [5,6]. For example, the study conducted by Dorshorst has shown a complex recombination structure region on chromosome 20 of silky fowl, and the END3 gene in this region was found to affect the excessive deposition of melanin [7]. The CNV of the KIT gene has been significantly associated with the dominant traits of coat color in pigs [8]. Another study on the Qinchuan cattle has reported the body height to be affected by the GBP4 gene CNV [9]. Several literature have proved that CNVs are crucial for studying the differences in growth traits of domestic animal, providing an important reference value for breeding work.
Yak is an animal unique to the plateau region [10], that is mainly distributed in the cold plateau zone. The yaks in China account for more than 95% of the world. Yaks provide multiple resources such as meat, milk, wool, and transportation to the herdsmen [11]. This experiment used the Ashidan yak, which shows excellent production performance. Ashidan yak is hornless and therefore suitable for barn feeding breeding in the frigid plateau area. As a new breed, the Ashidan yak is beneficial for developing the ecology and economy of locals. So far, several studies have established that CNV can influence economic traits [12,13,14], and the application of CNV has great potential in livestock genetic breeding for improving the economic benefits.
The SOX6 gene belonging to a transcription factor of the SRY (sex-determining region Y) family, was originally isolated from the testis of adult mice [15,16]. Over the last two decades, the literature has enlightened us about the SOX6 gene, indicating that the SOX6 gene might affect the nervous system [17], embryonic development [18], and sex determination [19]. In addition, the SOX6 gene is also involved in regulating the specification of the muscle fiber type in mammals [20]. Studies on the effects of SOX6 on the muscle and bone have identified the transcription factors in the SOX family to exert at point by targeting organs, showing interactions between the transcription factors. For instance, the proliferation and differentiation of the chondrocytes were regulated by the interaction of SOX5 and SOX6 with SOX9 [21]. SOX5 and SOX6 were SHOX interacting proteins, and their interaction can affect bone formation and development [22]. Based on Northern blot technology, the SOX6 gene was detected to be the highest in the skeletal muscle tissues of adult mice, indicating that SOX6 might be involved in maintaining the muscles [23].
Our previous study has utilized the whole-genome sequencing technology for detecting multiple genes located in the CNV region, including the SOX6 gene [24]. These CNV regions were located in the quantitative trait locus (QTLs) and were closely related to the growth traits in yaks. So far, there has been no report on the CNV of the SOX6 gene in yaks. Therefore, based on preliminary sequencing data, we would like to explore the correlation between the SOX6-CNV and growth traits in Ashidan yak. Moreover, we expect it could provide data support for the genetic improvement of yak breeding.
2. Materials and Methods
2.1. Animal Welfare
The Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS (No. LIHPS-CAAS-2017-115) has approved all experiments in this study. The body size traits and the blood samples of the yaks were assessed according to the Guidelines for the Care and Use of Laboratory Animals.
2.2. Body Size Traits and the Blood Samples Collection
All the blood samples of 311 healthy female Ashidan yaks were collected from the Datong Cattle Farm, Qinghai Province, China; each blood sample was 4 ml. The body indices including body weight, withers height, body length, and chest girth were measured for 311 yaks aged 6, 12, 18, and 30 months, respectively. The method for the standard measurement can be based on the study by Gilbert [25]. Seven tissues of three 3-year-old yaks were sampled for analyzing the SOX6 gene expression, for the tissues of the heart, liver, spleen, lungs, kidney, skeletal muscle, and adipose tissue. It is necessary to select yak raised under the same conditions and similar in body conformation. The three yaks were subjected to electric shock in this study to reduce the pain before death.
2.3. Isolation and Identification of Genomic the DNA and RNA
The genomic DNA was extracted from the blood using the EasyPure Blood Genomic DNA Kit. The RNA was isolated from the seven tissues of 3 yaks using the Trizol reagent and the RNA was purified using the RNase-free DNase based on the instructions. The purity and quality of DNA and RNA were determined using the Thermo Scientific NanoDrop 2000C and 1.2% agarose gel electrophoresis. Total RNAs were converted to cDNA using PrimeScript™ Reagent Kit and gDNA Eraser. DNA and cDNA samples were preserved at −20 °C. The main test reagent equipment and manufacturers in this experiment are shown in Table 1.

Table 1.
Main test reagent equipment and manufacturers.
2.4. Information of Candidate Gene
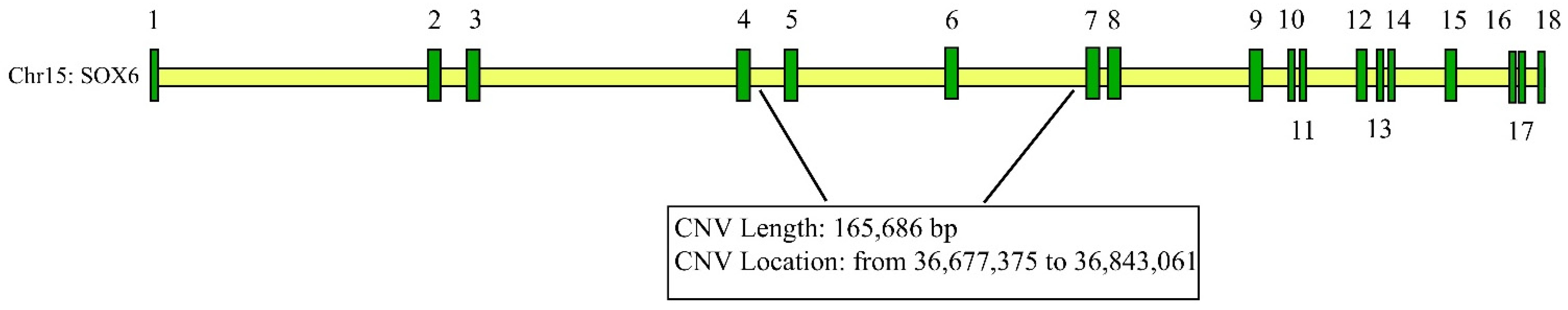
The CNV (chr15: 36,677,375 to 36,843,061) of SOX6 (AC_000172.1) is located in intron 4–6 (Figure 1).
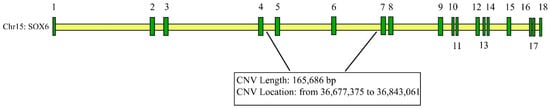
Figure 1.
Information on CNVs of SOX6 genes (the numbers from 1 to 18 denote the exons).
2.5. Primer Design
The gene sequence was queried using National Center for Biotechnology Information (NCBI). The primers were designed by the Primer-BLAST online software for analyzing the CNV and gene expression. Meanwhile, the primers were designed according to general principles [26]. For CNV analysis, the primers should be within the range of the SOX6-CNV region, and the basic transcription factor 3 (BTF3), which is known to be a universal transcription factor 3 was the reference gene in this study. To design primers for analyzing the gene expression according to the mRNA sequence of the SOX6 gene, the beta-actin (β-Actin) gene was selected as the reference gene. Details of the primer pair sequences information are shown below (Table 2). The primers and the optimal temperature of the primers were tested using the polymerase chain reaction (PCR) and 1% agarose gel electrophoresis.

Table 2.
Details of primers.
2.6. Copy Number Variation Identification and Gene Expression
Studies of CNV have generally employed qPCR technology, which is known for its validity and convenience [27]. The qPCR was performed on the LightCycler® 96 Instrument to detect the CNV-SOX6. A 20 μL reaction system was selected for the experiment, including 1 μL DNA/cDNA, 10 μL SYBR Premix Ex Taq II, 1 μL forward primers, 1 μL reverse primers, and 7 μL ddH2O. The qPCR procedure as shown below: 95 °C pre-denaturation for 30 s, 45 cycles involved 95 °C for 5 s, 59 °C for 30 s, after cycling, 5 s at 95 °C, 60 s at 65 °C, and finally at 95 °C continuously. All the experiments were replicated three times to ensure the accuracy of the experiment, and the final data were presented as mean ± standard deviation (SD).
2.7. CNV Correlation Analysis and Expression Profiling
The final value were calculated according to the formula:
where ΔCt = Cttarget gene − Ctreference gene, ΔΔCt = ΔCttest − ΔCtcontrol, and all the analysis data were standardized [28]. The expression profiling of the SOX6 gene was plotted using the GraphPad Prism 8.0 software. The relevance between the SOX6-CNV and four phenotypes was detected using the SPSS 26.0 software. The statistical method was ANOVA (analysis of variance) and the non-parametric test. Before ANOVA, the homogeneity test (Table S1) and normality test (Table S2) were carried out for each character in the different CNV regions. The general linear model approach was chosen considering the uncertain factors influencing the phenotypic value, including age, genetic effects, and environment. The correlation between the CNV and four growth traits was analyzed using the model:
where Yj represents the observed value of growth traits; CNVj represents the SOX6-CNV type effect; μ represents the total mean value of each character; ej means random residual and j represents jth CNV type.
2 × 2−ΔΔCt.
Yj = μ + CNVj + ej.
3. Results
3.1. Expression Profiling of SOX6 Gene and Distribution of Different CNV Types in Yaks
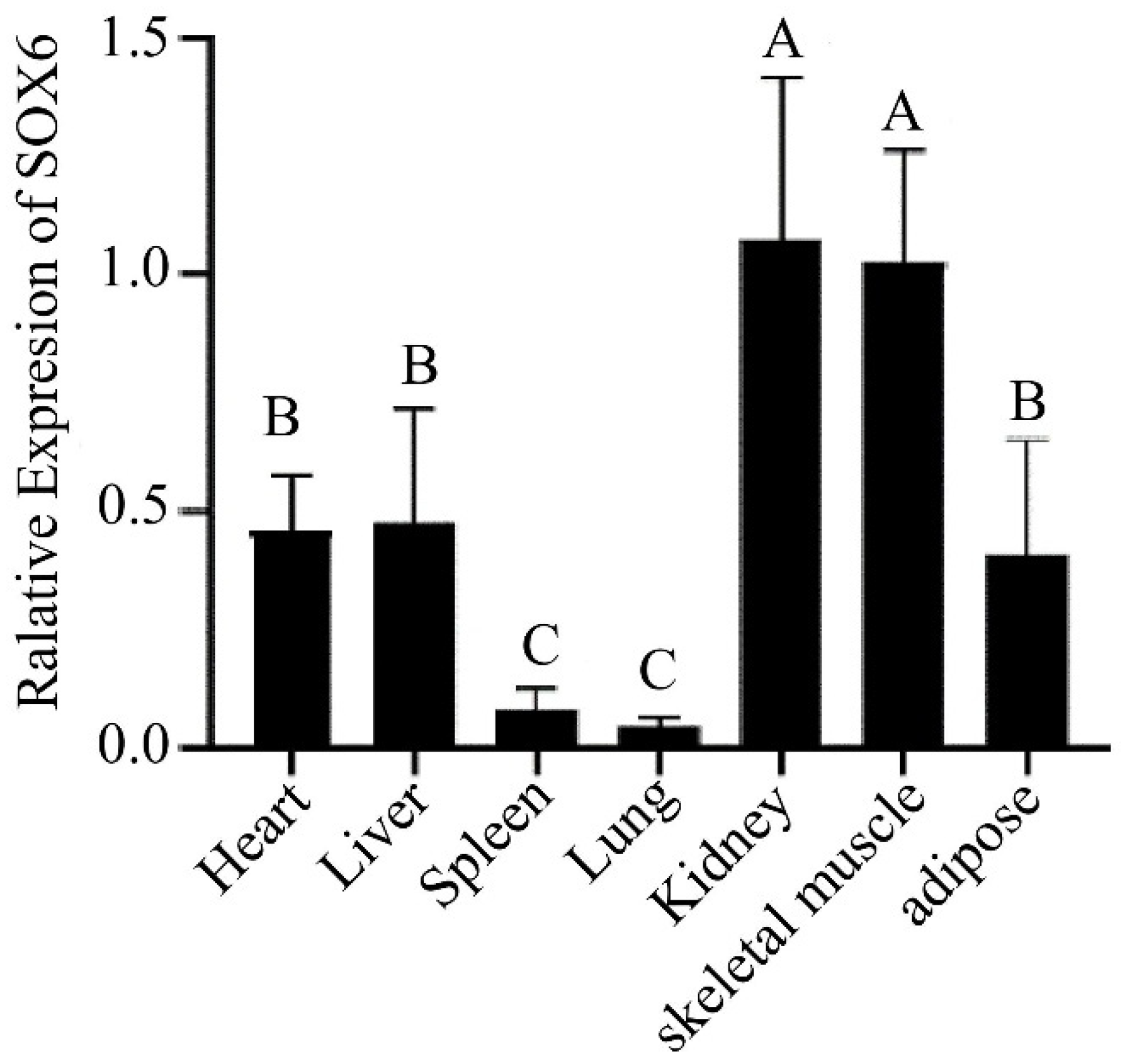
The SOX6 gene as a transcription factor regulates physiological functions such as regulating chondrogenesis, nervous system development, erythropoiesis, and other processes. The expression of the SOX6 gene was detected in seven tissues of three yaks using qPCR, and the results are displayed in Figure 2. The SOX6 gene was detected in all the tissues and the most abundant expression was found in the kidney and skeletal muscle tissues while low expressions were evident in the spleen and lung tissues. Furthermore, the experimental data were processed through the cycle threshold value for obtaining the type and frequency of the SOX6-CNV. The data were divided into three categories by 2 × 2−ΔΔCt < 1 (Loss), 1 ≤ 2 × 2−ΔΔCt ≤ 2 (Normal), 2 × 2−ΔΔCt > 2 (Gain) [29]. The number of SOX6-CNV in Loss, Normal and Gain are 17, 182, and 112. By calculating the proportions of the three categories of data in 311 yaks, we found that the CNV distribution of SOX6 gene was different among the three types of yaks. The loss type CNV frequency of the SOX6 gene was found to be 5%, while that of the normal type CNV frequency was 59% and the gain type CNV frequency was 36%.
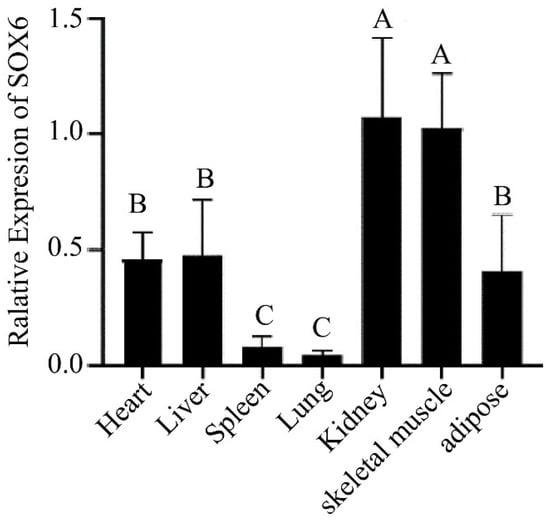
Figure 2.
Tissues expression of SOX6 in the Ashidan yak. Different capital letters (A, B, C) represent the extremely significant differences (p < 0.01).
3.2. Correlation between the SOX6-CNV and Four Growth Traits at Different Ages of 311 Yaks
A multitude of studies has confirmed that CNV extensively affects the growth traits of animals. After the homogeneity of the variance test, the SOX6-CNV and growth traits were tested to verify the relative using ANOVA. The experimental evidence in Table 3 showed the correlation analysis between the different types of SOX6-CNV and four economic traits of Ashidan yak. The chest girth of 6-month yaks (p < 0.05), withers height of 6-month yaks (p < 0.05), withers height of 18-month yaks (p < 0.05), chest girth of 30-month yaks (p < 0.05) were significantly correlated with SOX6-CNV. Subsequently, multiple comparisons were made based on the ANOVA analysis of variance for evaluating the differences between the means. The different CNV types of SOX6-CNV were found to significantly affect the growth traits. However, the CNV advantage types of the withers height were not uniform, the loss type has a better withers height at 6 months of age but the advantage was not obvious. Moreover, the withers height of gain type was significantly superior to the other types at 18 months of age. Meanwhile, the chest girth of the normal-type yak was generally better than that of the other types. Combining the data in Table 3, the normal types of individuals showed better growth traits. The results indicated that the SOX6 gene variable region affects some important growth traits of yaks.

Table 3.
ANOVA of the correlation between SOX6-CNV and four growth traits in 311 Ashidan yaks.
4. Discussion
CNV was originally applied for studying the rod chromosomes of Drosophila [30]. Subsequently, researchers have studied CNVs for exploring human genetic diseases and livestock breeding [31,32]. With the in-depth study of genomics, CNV has been at the center of much attention, and researchers have made significant progress in this field. Due to the crucial nature of the economic traits, more attention needs to be paid to breeding individuals of excellent characteristics using molecular breeding. So far, previous studies have revealed a correlation between the CNV and some traits of animals. CNV has been studied in livestock such as chickens [33], pigs [34], sheep [2], cattle [29], etc. In addition, there was a significant correlation between the HPGDS-CNV and body weight aged 12, 18, and 30 months Ashidan yaks, and the body length of 18, 30-month-old yaks has been illustrated in a previous study of our group. The HPGDS gene can be employed as a candidate gene for MAS in yak breeding [35].
SOX6 was originally isolated from the testis of the adult mice [15,16], and was known to perform the function by immune-stimulating gene expression of the type II collagen and aggrecan during the proliferating phase [36]. Prior studies have noted the importance of this gene in invertebrate growth and development of invertebrates which can regulate mouse spermatogenesis and participate in controlling neural differentiation [37,38,39]. In addition, there were several reports that the SOX6 gene was also closely related to muscle development, the gene expression was most abundant in the skeletal muscle of the adult mice [40,41,42,43,44]. Further research found that SOX6 takes part in regulating muscle fiber types and skeletal muscle growth and development [45,46]. In summary, numerous previous studies described a connection between the SOX6 gene and the type transition of the skeletal muscle fiber. Therefore, it is meaningful for us to explore the effects of SOX6 on the growth traits in yaks.
Reviewing our study, the gene expression levels and SOX6-CNV were detected by qPCR. One interesting finding was that the SOX6 gene was expressed in all seven tissues. Figure 1 shows that the SOX6 gene was adequately expressed in the kidney and heart tissues. A possible explanation for this might be that the yaks possess a more efficient cardiopulmonary function and a metabolic system in the high altitude and low oxygen environment [47,48]. The higher expression level of the SOX6 gene was found in the skeletal muscle tissues. Combined with the previously mentioned finding, the SOX6 was found to be related to muscle development. A comparison of the findings with the expression profile of SOX6 in the human tissues on NCBI confirmed the reliability of our data. Results on CNV of the SOX6 gene displayed that SOX6-CNV was statistically significantly related to the withers height and the chest girth of the 6-month yaks, the withers height of the 18-month yaks, the chest girth of the 30-month yaks. These results might be explained by the fact that yaks need to adapt to the plateau environment. The yak calves were weaned until 6-month old, to ensure that they can get abundant nutrients from breast milk during lactation. The body composition changes corresponded during this period. To adapt to the plateau, if their muscles and fat were not fully developed, the yaks would adapt to the environment of low oxygen mainly by heart. The heart developed rapidly during this period, and the thoracic cavity gradually became larger and deeper. When the yaks were 18-mouths old, the development of the body was speculated to the abundant pasture during this period and the increasing feed intake of the yak. The nutrient intake was greatly helpful for the daily weight gain of the yak during the growth period, muscle and fat growth development [49]. In addition, the chest girth of 30-month-old yaks was found to be significantly correlated with SOX6-CNV, which might be the reason why SOX6 gene expression in the chest muscle was higher than that in the leg muscle [50]. The above results indicated that the SOX6 gene might affect the growth and development of yaks, which might be related to the unique adaptability of the yak to the plateau.
The development of animal husbandry in the plateau areas is dispensable for the yaks and is necessary for selecting and breeding yaks. However, the slow growth of yaks has been a major problem in yak breeding. The withers height and chest girth are important growth traits for improving the defect of yak performance and economic benefits, of course, this requires molecular breeding technology. This study has identified that the SOX6-CNV is associated with the phenotype of the yaks and has a significant correlation with the chest girth at 6-mouth age and 30-mouth age and withers height at 6-mouth age and 18-mouth age of the Ashidan yak. According to the experimental data, the CNV advantage types of chest girth and withers height were not consistent at all ages, but there were tendencies for the individuals with the normal type to have a higher value on the growth traits. In addition, it is not practical to breed the yak only at a certain age since it does not consider the whole growth period of the yaks. Comprehensively, in this study, the normal type of CNV should have better advantages. It is possible that the SOX6 gene can be a reference gene for yak breeding in the future. The research can not only provide a reference for the genetic improvement of yaks but also complement the research deficiency of the SOX6 gene in the growth traits of yaks. Moreover, further research on different yak breeds should be conducted, especially other hybrid improved breeds with outstanding growth performance. Moreover, the mechanism of the SOX6-CNV affecting the growth traits needs further investigation.
5. Conclusions
The research aimed to assess if the SOX6 gene influences the growth traits of yaks. This study has examined the impact of the CNV of the SOX6 gene in yaks, and the SOX6-CNV was found to be significantly correlated to the chest girth and the withers height of 6-month yak (p < 0.05), the withers height of 18-month yak (p < 0.05), and the chest girth of 30-month yak (p < 0.05). The expression of the SOX6 gene in the yak tissues was detected at the mRNA level, and a high expression of the SOX6 gene was found in the muscle and kidney tissues. The evidence from this study suggests that the SOX6 gene might influence the growth and development of the yak muscles, which provided a reference for studying the SOX6 gene and can be considered as a candidate gene by molecular technique to assist in yak breeding.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ani12223074/s1. Table S1: The homogeneity of variance test of each trait. Table S2: The test of normality of each trait.
Author Contributions
Conceptualization, X.L. and C.H.; methodology, X.L. and M.L.; software, X.L.; validation, X.L. and R.D.; formal analysis, X.L.; investigation, X.L.; resources, X.L., C.H., R.D. and R.D.; data curation, X.L. and C.H.; writing—original draft preparation, X.L.; writing—review and editing, X.L. and C.L.; visualization, P.Y., C.L., X.W., M.C., P.B., J.P., X.G. and X.M.; supervision, X.W., M.C., P.B., J.P., X.G. and X.M.; project administration, C.L.; funding acquisition, C.L., X.W., M.C., P.B., J.P., X.G. and X.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by State Key R & D program (2021YFD1600200); Modern beef yak industry technology system (MATS-Beef Cattle System, CARS-37); Gansu basic research innovation group project (20JR5RA580); Yak resources and breeding innovation project of Chinese Academy of Agricultural Sciences (25-LIHPS-01).
Institutional Review Board Statement
The procedures and protocols of all experiments in this study have been approved by the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS (Grant Number is No. LIHPS-CAAS-2017-115).
Informed Consent Statement
The animal samples were collected approved by Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS (approval NO. LIHPS-CAAS-2017-115). The body size traits and the blood samples of the yaks were assessed according to the Guidelines for the Care and Use of Laboratory Animals, samples were also taken with the consent of the farmer and related personnel.
Data Availability Statement
The study did not report any data.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Feuk, L.; Marshall, C.R.; Scherer, W.S.W. Structural variants: Changing the landscape of chromosomes and design of disease studies. Hum. Mol. Genet. 2006, 15, R57. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.Y.; Li, L.J.; Zhang, Z.J.; Wang, E.Y.; Huang, Y.Z. Copy number variation of MYLK4 gene and its growth traits of Capra hircus (goat). Anim. Biotechnol. 2019, 31, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Korbel, J.O.; Urban, A.E.; Affourtit, J.P.; Godwin, B.; Grubert, F.; Simons, J.F.; Kim, P.M.; Palejev, D.; Carriero, N.J.; Du, L. Paired-End Mapping Reveals Extensive Structural Variation in the Human Genome. Science 2007, 318, 420–426. [Google Scholar] [CrossRef]
- Rogers, R.L.; Cridland, J.M.; Shao, L.; Hu, T.T.; Peter, A.; Thornton, K.R. Landscape of standing variation for tandem duplications in Drosophila yakuba and Drosophila simulans. Mol. Biol. Evol. 2014, 37, 1750–1766. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P.; Choi, J.; Basu, U.; Sumnerthomson, J.M.; Yan, M.; Liao, X.; Moore, S.S. Whole genome resequencing of Black Angus and Holstein cattle for SNP and CNV discovery. BMC Genom. 2011, 12, 559. [Google Scholar] [CrossRef] [PubMed]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B. The variome concept: Focus on CNVariome. Mol. Cytogenet. 2019, 12, 52. [Google Scholar] [CrossRef]
- Dorshorst, B.; Molin, A.M.; Rubin, C.J.; Johansson, A.M.; StrMstedt, L.; Pham, M.H.; Chen, C.F.; Hallb, F.K.; Shwell, C.A.; Andersson, L. A Complex Genomic Rearrangement Involving the Endothelin 3 Locus Causes Dermal Hyperpigmentation in the Chicken. PLoS Genet. 2011, 7, e1002412. [Google Scholar] [CrossRef]
- Seo, B.Y.; Park, E.W.; Ahn, S.J.; Lee, S.H.; Jeon, J.T. An accurate method for quantifying and analyzing copy number variation in porcine KIT by an oligonucleotide ligation assay. BMC Genet. 2007, 8, 81. [Google Scholar] [CrossRef]
- Xiong, Y.C.; Cao, X.K.; He, H.; Song, L.F.; Huang, C.; Dang, L.P.; Zhang, J.Y.; Lei, C.Z.; Cheng, H.; Qi, X.L.; et al. The Detection of GBP4 Gene Copy Number Variation and Its Effect on Five Bovine Growth Traits. China Cattle Sci. 2016, 42, 9–12. [Google Scholar]
- Wright, D.; Boije, H.; Meadows, J.R.S.; Bed’hom, B.; Gourichon, D.; Vieaud, A.; Tixier-Boichard, M.; Rubin, C.J.; Imsland, F. Copy Number Variation in Intron 1 of SOX5 Causes the Pea-comb Phenotype in Chickens. PLoS Genet. 2009, 5, e1000512. [Google Scholar] [CrossRef]
- Ding, L.; Wang, Y.; Kreuzer, M.; Guo, X.; Mi, J.; Gou, Y.; Shang, Z.; Zhang, Y.; Zhou, J.; Wang, H.; et al. Seasonal variations in the fatty acid profile of milk from yaks grazing on the Qinghai-Tibetan plateau. J. Dairy Res. 2013, 80, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, L.Z.; Shi, T.; Zhou, Y.; Cai, H.F. Copy number variations of MICAL-L2 shaping gene expression contribute to different phenotypes of cattle. Mamm. Genome 2013, 24, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.M.; Li, Z.; He, H.; Song, C.C.; Zhang, Z.J. Associations of GBP2 gene copy number variations with growth traits and transcriptional expression in Chinese cattle. Gene 2018, 647, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Henrique, D.; Junior, G.; Cesar, A.; Freua, M.C.; Gomes, R.C.; Da, L.; Leme, P.R.; Fukumasu, H.; Carvalho, M.E.; Ventura, R.V. Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle. J. Appl. Genet. 2016, 57, 495–504. [Google Scholar]
- Kondoh, H.; Kamachi, Y. SOX-partner code for cell specification: Regulatory target selection and underlying molecular mechanisms. Int. J. Biochem. Cell Biol. 2009, 42, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef]
- Hamada-Kanazawa, M.; Ishikawa, K.; Ogawa, D.; Kanai, M.; Kawai, Y.; Narahara, M.; Miyake, M. Suppression of SOX6 in P19 cells leads to failure of neuronal differentiation by retinoic acid and induces retinoic acid-dependent apoptosis. FEBS Lett. 2004, 577, 60–66. [Google Scholar] [CrossRef]
- Lefebvre, V.; Crombrugghe, B.D.; Behringer, R.R. The transcription factors L-SOX5 and SOX6 are essential for cartilage formation. Dev. Cell 2001, 1, 277–290. [Google Scholar]
- Lefebvre, V. The SOXD transcription factors—SOX5, SOX6, and SOX13—Are key cell fate modulators. Int. J. Biochem. Cell Biol. 2010, 42, 429–432. [Google Scholar] [CrossRef]
- Jackson, H.E.; Ono, Y.; Wang, X.; Elworthy, S.; Cunliffe, V.T.; Ingham, P.W. The role of SOX6 in zebrafish muscle fiber type specification. Skelet. Muscle 2015, 5, 2. [Google Scholar] [CrossRef]
- Sun, D.M. Studies on the Transcriptional Regulation Mechanisms of Transcription Factors Required for Skeletal Development; Northeast Normal University: Changchun, China, 2009. [Google Scholar]
- Aza-Carmona, M.; Shears, D.J.; Yuste-Checa, P.; Barca-Tierno, V.; Hisado-Oliva, A.; Belinchón, A.; Benito-Sanz, S.; Rodríguez, J.I.; Argente, J.; Campos-Barros, A.; et al. SHOX interacts with the chondrogenic transcription factors SOX5 and SOX6 to activate the aggrecan enhancer. Hum. Mol. Genet. 2011, 20, 1547–1559. [Google Scholar] [CrossRef] [PubMed]
- Klewer, H.S.E.; Samson, R.A. SOX6 is a candidate gene for p100H myopathy, heart block, and sudden neonatal death. Proc. Natl. Acad. Sci. USA 2000, 97, 4180–4185. [Google Scholar]
- Jia, C.; Wang, H.; Li, C.; Wu, X.; Yan, P. Genome-wide detection of copy number variations in polled yak using the Illumina BovineHD BeadChip. BMC Genom. 2019, 20, 376. [Google Scholar] [CrossRef]
- Gilbert, R.P.; Bailey, D.R.; Shannon, N.H. Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets. J. Anim. Sci. 1993, 71, 1712. [Google Scholar] [CrossRef]
- Edwards, E.; Saunders, N.; Logan, J.; Sails, A.D.; Ad, S. Real-Time PCR: Current Technology and Applications; Caister Academic Press: Poole, UK, 2009; p. 284. [Google Scholar]
- Ali, S.; Srivastava, A.K.; Chopra, R.; Aggarwal, S.; Garg, V.K.; Bhattacharya, S.N.; Bamezai, R. IL12B SNPs and copy number variation in IL23R gene associated with susceptibility to leprosy. J. Med. Genet. 2013, 50, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Cheong, H.S.; Kim, L.H.; Namgung, S.; Park, T.J.; Chun, J.Y.; Kim, J.Y.; Pasaje, C.; Jin, S.L.; Shin, H.D. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genom. 2010, 11, 232. [Google Scholar] [CrossRef]
- Liu, M.; Li, B.; Shi, T.; Huang, Y.; Liu, G.E.; Lan, X.; Lei, C.; Chen, H. Copy number variation of bovine SHH gene is associated with body conformation traits in Chinese beef cattle. J. Appl. Genet. 2019, 60, 199–207. [Google Scholar] [CrossRef]
- Knudsen, O. Studies on spermatocytogenesis in the bull. Int. J. Fertil. 1958, 3, 389–403. [Google Scholar]
- Yang, X.; Song, Z.; Wu, C.; Wang, W.; Li, G.; Zhang, W.; Wu, L.; Lu, K. Constructing a database for the relations between CNV and human genetic diseases via systematic text mining. BMC Bioinform. [Electron. Resour.] 2018, 19 (Suppl. 19), 528. [Google Scholar] [CrossRef]
- Ma, Y.L.; Wen, Y.F.; Cao, X.K.; Cheng, J.; Chen, H. Copy number variation (CNV) in the IGF1R gene across four cattle breeds and its association with economic traits. Arch. Anim. Breed. 2019, 62, 171–179. [Google Scholar] [CrossRef]
- Jing, Z.; Wang, X.; Cheng, Y.; Wei, C.; Hou, D.; Li, T.; Li, W.; Han, R.; Li, H.; Sun, G. Detection of CNV in the SH3RF2 gene and its effects on growth and carcass traits in chickens. BMC Genet. 2020, 21, 22. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.G.; Zhang, Y.B.; Yan, H.; Zhang, L.C.; Hou, X.H.; Liu, X.; Wang, L.X. Identification of Candidate Genes for Porcine Bone Rate Traitsby Genome-wide Association of Copy Number Variation. Acta Vet. Zootech. Sin. 2019, 50, 7. [Google Scholar]
- Huang, C.; Ge, F.; Ren, W.; Zhang, Y.; Liang, C. Copy Number Variation of the HPGDS Gene in the Ashidan yak and Its Associations with Growth Traits. Gene 2020, 772, 145382. [Google Scholar] [CrossRef]
- Ikeda, T.; Kawaguchi, H.; Kamekura, S.; Kou, I.; Hoshi, K.; Nakamura, K.; Ikegawa, S.; Chung, U. Combination of SOX5, SOX6 and SOX9 (the SOX trio) provides signals suffucient for introduction of permanent cartilage. Arthritis Rheum. 2004, 50, 3561–3573. [Google Scholar] [CrossRef] [PubMed]
- Frances, C.; Edwina, W.; Paul, D.; Peter, K.; Alan, A. The Sry-related HMG box-containing gene SOX6 is expressed in the adult testis and developing nervous system of the mouse. Nucleic Acids Res. 1995, 23, 3365. [Google Scholar]
- Cantone, M.; Küspert, M.; Reiprich, S.; Lai, X.; Eberhardt, M.; Gttle, P.; Beyer, F.; Azim, K.; Küry, P.; Wegner, M. A gene regulatory architecture that controls region-independent dynamics of oligodendrocyte differentiation. Glia 2019, 67, 825–843. [Google Scholar] [CrossRef]
- Zhang, L.; Xue, Z.; Yan, J.; Wang, J.; Liu, Q.; Jiang, H. LncRNA Riken-201 and Riken-203 modulates neural development by regulating the SOX6 through sequestering miRNAs. Cell Prolif. 2019, 52, e12573. [Google Scholar] [CrossRef]
- Sluijter, J.; Mil, A.V.; Vliet, P.V.; Metz, C.; Liu, J.; Doeven Da Ns, P.A.; Goumans, M.J. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arter. Thromb. Vasc. Biol 2010, 30, 859–868. [Google Scholar] [CrossRef]
- Lübbert, M.; Jones, P.A. Epigenetic Regulation of Globin Genes and Disturbances in Hemoglobinopathies; Springer: Berlin/Heidelberg, Germany, 2014; pp. 89–106. [Google Scholar] [CrossRef]
- An, C.I.; Dong, Y.; Nobuko, H. Genome-wide mapping of SOX6 binding sites in skeletal muscle reveals both direct and indirect regulation of muscle terminal differentiation by SOX6. BMC Dev. Biol. 2011, 11, 59. [Google Scholar] [CrossRef]
- Rooij, E.V.; Quiat, D.; Johnson, B.A.; Sutherland, L.B.; Qi, X.; Richardson, J.A.; Kelm, R.J.; Olson, E.N. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 2009, 17, 662–673. [Google Scholar] [CrossRef]
- Smits, P. SOX5 and SOX6 are needed to develop and maintain source, columnar, and hypertrophic chondrocytes in the cartilage growth plate. J. Cell Biol. 2004, 164, 747–758. [Google Scholar] [CrossRef] [PubMed]
- Quiat, D.; Voelker, K.A.; Pei, J.; Grishin, N.V.; Grange, R.W.; Bassel-Duby, R.; Olson, E.N. Concerted regulation of myofiber-specific gene expression and muscle performance by the transcriptional repressor SOX6. Proc. Natl. Acad. Sci. USA 2011, 108, 10196–10201. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y. MicroRNA-499-5p Regulates the Formation of Porcine Slow Myofibers by Targeting SOX6. Master’s Thesis, Sichuan Agricultural University, Ya’an, China, 2017. [Google Scholar]
- Wiener, G.; Han, J.; Long, R. The yak. Rap Publ. 2011, 44, 57–58. [Google Scholar]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 8. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.F. Study on Main Factors Influencing the Growth of Yak under the Condition of Grazing Grass at Different Growing Times during Warm Season. China Dairy Cattle 2021, 5–8. [Google Scholar] [CrossRef]
- Lin, X.R. Copy Number Variation of the SOX6 Affects Chicken Growth Traits. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2016. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).