Transcriptome Sequencing Analysis of circRNA in Skeletal Muscle between Fast- and Slow-Growing Chickens at Embryonic Stages
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissues
2.2. Construction of cDNA Library
2.3. Bioinformatics Analysis for the Sequencing Data
2.4. Construction of circRNA–miRNA Interaction Network
2.5. Validation for Differentially Expressed circRNAs
2.6. Statistical Analysis
3. Results
3.1. Statistics of Chicken Egg Weight and Embryo Bodyweight
3.2. Summary of Sequencing Data
3.3. Statistics of Differentially Expressed circRNAs
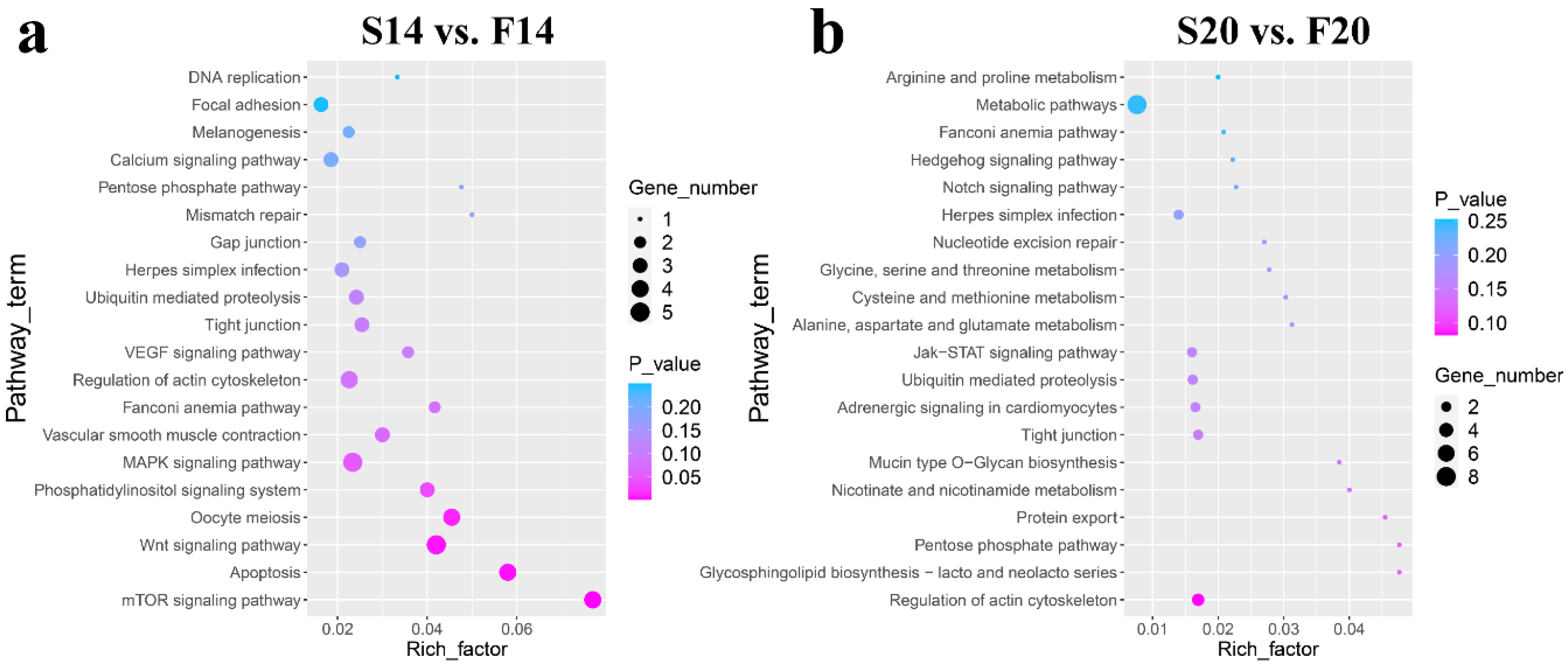
3.4. Functional Analysis of the Host Genes of Differentially Expressed circRNAs
3.5. CircRNA–miRNA Interaction Network
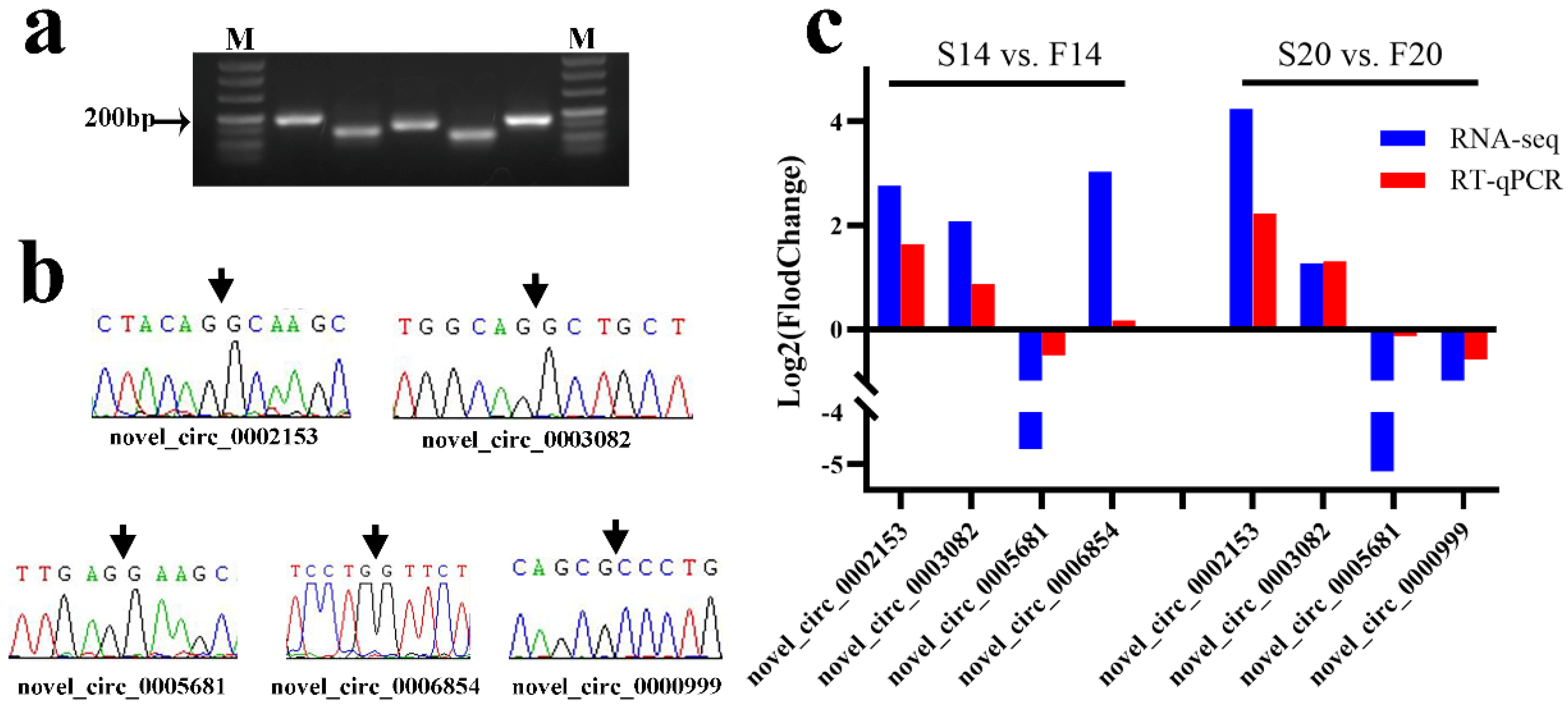
3.6. Verification of circRNA Sequencing Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, W.; Lv, Y.T.; Zhang, H.X.; Ruan, D.; Wang, S.; Lin, Y.C. Developmental specificity in skeletal muscle of late-term avian embryos and its potential manipulation. Poult. Sci. 2013, 92, 2754–2764. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhang, S.; Xu, Z.; Gao, J.; Mishra, S.K.; Zhu, Q.; Zhao, X.; Wang, Y.; Yin, H.; Fan, X.; et al. MiRNA Profiling in Pectoral Muscle Throughout Pre- to Post-Natal Stages of Chicken Development. Front. Genet. 2020, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, C.; Sun, H.; Wang, J.; Liang, Y.; Wang, Y.; Wong, G. The bioinformatics toolbox for circRNA discovery and analysis. Brief. Bioinform. 2021, 22, 1706–1728. [Google Scholar] [CrossRef] [PubMed]
- Diener, T.O. Potato spindle tuber “virus”. IV. A replicating, low molecular weight RNA. Virology 1971, 45, 411–428. [Google Scholar] [CrossRef]
- Sanger, H.L.; Klotz, G.; Riesner, D.; Gross, H.J.; Kleinschmidt, A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA 1976, 73, 3852–3856. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Liu, R.; Liu, X.; Bai, X.; Xiao, C.; Dong, Y. Identification and Characterization of circRNA in Longissimus Dorsi of Different Breeds of Cattle. Front. Genet. 2020, 11, 565085. [Google Scholar] [CrossRef]
- Jiao, H.; Zhao, Y.; Zhou, Z.; Li, W.; Li, B.; Gu, G.; Luo, Y.; Shuai, X.; Fan, C.; Wu, L.; et al. Identifying Circular RNAs in HepG2 Expressing Genotype IV Swine Hepatitis E Virus ORF3 Via Whole Genome Sequencing. Cell Transplant. 2021, 30, 9636897211055042. [Google Scholar] [CrossRef]
- Quan, G.; Li, J. Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian. Res. 2018, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Das, A.; Das, D.; Abdelmohsen, K.; Panda, A.C. Circular RNAs in myogenesis. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194372. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Lian, C.; Chen, G.; Zou, P.; Qin, B.G. CircRNA FUT10 regulates the regenerative potential of aged skeletal muscle stem cells by targeting HOXA9. Aging 2021, 13, 17428–17441. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Shyamal, S.; Sinha, T.; Mishra, S.S.; Panda, A.C. Identification of Potential circRNA-microRNA-mRNA Regulatory Network in Skeletal Muscle. Front. Mol. Biosci. 2021, 8, 762185. [Google Scholar] [CrossRef] [PubMed]
- Legnini, I.; Di Timoteo, G.; Rossi, F.; Morlando, M.; Briganti, F.; Sthandier, O.; Fatica, A.; Santini, T.; Andronache, A.; Wade, M.; et al. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell 2017, 66, 22–37.e29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.X.; Zhang, T.; Wei, Y.; Ding, F.X.; Zhang, L.; Wang, J.Y. Functional identification of an exon 1 substitution in the myostatin gene and its expression in breast and leg muscle of the Bian chicken. Br. Poult. Sci. 2015, 56, 639–644. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, J.; Zhao, F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018, 19, 803–810. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Doran, T.J.; Cooper, C.A.; Jenkins, K.A.; Tizard, M.L.V. Advances in genetic engineering of the avian genome: “Realising the promise”. Transgenic Res. 2016, 25, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Mugo, P.W. Assessment of Availability and Use of Information and Communication Technology in Broiler Marketing: The Case of Peri-Urban Broiler Farmers in Njiru District-Nairobi County. Ph.D. Thesis, University of Nairobi Kenya, Nairobi, Kenya, 2012. [Google Scholar]
- Soglia, F.; Petracci, M.; Davoli, R.; Zappaterra, M. A critical review of the mechanisms involved in the occurrence of growth-related abnormalities affecting broiler chicken breast muscles. Poult. Sci. 2021, 100, 101180. [Google Scholar] [CrossRef] [PubMed]
- Tijare, V.V.; Yang, F.L.; Kuttappan, V.A.; Alvarado, C.Z.; Coon, C.N.; Owens, C.M. Meat quality of broiler breast fillets with white striping and woody breast muscle myopathies. Poult. Sci. 2016, 95, 2167–2173. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Huang, W.; Wang, Z.; Chen, Y.; Cai, D.; Nie, Q. circTAF8 Regulates Myoblast Development and Associated Carcass Traits in Chicken. Front. Genet. 2021, 12, 743757. [Google Scholar] [CrossRef]
- Zhao, J.; Zhao, X.; Shen, X.; Zhang, Y.; Zhang, Y.; Ye, L.; Li, D.; Zhu, Q.; Yin, H. CircCCDC91 regulates chicken skeletal muscle development by sponging miR-15 family via activating IGF1-PI3K/AKT signaling pathway. Poult. Sci. 2022, 101, 101803. [Google Scholar] [CrossRef]
- Yin, H.; Shen, X.; Zhao, J.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Wei, Y.; Wang, Y.; et al. Circular RNA CircFAM188B Encodes a Protein That Regulates Proliferation and Differentiation of Chicken Skeletal Muscle Satellite Cells. Front. Cell Dev. Biol. 2020, 8, 522588. [Google Scholar] [CrossRef]
- Sobolewska, A.; Elminowska-Wenda, G.; Bogucka, J.; Szpinda, M.; Walasik, K.; Bednarczyk, M.; Paruszewska-Achtel, M. Myogenesis--possibilities of its stimulation in chickens. Folia Biol. 2011, 59, 85–90. [Google Scholar] [CrossRef]
- Costa, M.L.; Jurberg, A.D.; Mermelstein, C. The Role of Embryonic Chick Muscle Cell Culture in the Study of Skeletal Myogenesis. Front. Physiol. 2021, 12, 668600. [Google Scholar] [CrossRef]
- Covarrubias, A.J.; Perrone, R.; Grozio, A.; Verdin, E. NAD(+) metabolism and its roles in cellular processes during ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 119–141. [Google Scholar] [CrossRef]
- Goody, M.F.; Henry, C.A. A need for NAD+ in muscle development, homeostasis, and aging. Skelet Muscle 2018, 8, 9. [Google Scholar] [CrossRef]
- Dao, T.; Green, A.E.; Kim, Y.A.; Bae, S.J.; Ha, K.T.; Gariani, K.; Lee, M.R.; Menzies, K.J.; Ryu, D. Sarcopenia and Muscle Aging: A Brief Overview. Endocrinol. Metab. 2020, 35, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Imai, S.; Guarente, L. NAD+ and sirtuins in aging and disease. Trends Cell Biol. 2014, 24, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Verdin, E. NAD⁺ in aging, metabolism, and neurodegeneration. Science 2015, 350, 1208–1213. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, Y.; Zhu, J.; Shi, Y.; Gu, W.; Kobe, B.; Ve, T.; DiAntonio, A.; Milbrandt, J. Nicotinic acid mononucleotide is an allosteric SARM1 inhibitor promoting axonal protection. Exp. Neurol. 2021, 345, 113842. [Google Scholar] [CrossRef] [PubMed]
- Pramono, A.A.; Rather, G.M.; Herman, H.; Lestari, K.; Bertino, J.R. NAD- and NADPH-Contributing Enzymes as Therapeutic Targets in Cancer: An Overview. Biomolecules 2020, 10, 358. [Google Scholar] [CrossRef]
- Pruller, J.; Figeac, N.; Zammit, P.S. DVL1 and DVL3 require nuclear localisation to regulate proliferation in human myoblasts. Sci. Rep. 2022, 12, 8388. [Google Scholar] [CrossRef]
- Song, Z.W.; Jin, C.L.; Ye, M.; Gao, C.Q.; Yan, H.C.; Wang, X.Q. Lysine inhibits apoptosis in satellite cells to govern skeletal muscle growth via the JAK2-STAT3 pathway. Food Funct. 2020, 11, 3941–3951. [Google Scholar] [CrossRef]
- Liu, J.; Jing, X.; Gan, L.; Sun, C. The JAK2/STAT3 signal pathway regulates the expression of genes related to skeletal muscle development and energy metabolism in mice and mouse skeletal muscle cells. Biosci. Biotechnol. Biochem. 2012, 76, 1866–1870. [Google Scholar] [CrossRef]
- Frontera, W.R.; Ochala, J. Skeletal muscle: A brief review of structure and function. Calcif. Tissue Int. 2015, 96, 183–195. [Google Scholar] [CrossRef]
- Thalacker-Mercer, A.; Riddle, E.; Barre, L. Protein and amino acids for skeletal muscle health in aging. Adv. Food Nutr. Res. 2020, 91, 29–64. [Google Scholar] [CrossRef]
- Pan, Z.; Yang, C.; Zhao, R.; Jiang, X.; Yu, C.; Li, Z. Characterization of lncRNA/circRNA-miRNA-mRNA network to reveal potential functional ceRNAs in the skeletal muscle of chicken. Front. Physiol. 2022, 13, 969854. [Google Scholar] [CrossRef] [PubMed]
- Kawano, T.; Inokuchi, J.; Eto, M.; Murata, M.; Kang, J.H. Activators and Inhibitors of Protein Kinase C (PKC): Their Applications in Clinical Trials. Pharmaceutics 2021, 13, 1748. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, H.A.; Bertrand, A.T.; Labib, S.; Mohamed-Uvaize, M.; Bolongo, P.M.; Wu, W.Y.; Bilińska, Z.T.; Bonne, G.; Akimenko, M.A.; Tesson, F. Protein Kinase C Alpha Cellular Distribution, Activity, and Proximity with Lamin A/C in Striated Muscle Laminopathies. Cells 2020, 9, 2388. [Google Scholar] [CrossRef] [PubMed]
- Pathak, P.; Blech-Hermoni, Y.; Subedi, K.; Mpamugo, J.; Obeng-Nyarko, C.; Ohman, R.; Molloy, I.; Kates, M.; Hale, J.; Stauffer, S.; et al. Myopathy associated LDB3 mutation causes Z-disc disassembly and protein aggregation through PKCα and TSC2-mTOR downregulation. Commun. Biol. 2021, 4, 355. [Google Scholar] [CrossRef]
- Kamada, R.; Kudoh, F.; Ito, S.; Tani, I.; Janairo, J.I.B.; Omichinski, J.G.; Sakaguchi, K. Metal-dependent Ser/Thr protein phosphatase PPM family: Evolution, structures, diseases and inhibitors. Pharmacol. Ther. 2020, 215, 107622. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef]
- Álvarez-Garcia, V.; Tawil, Y.; Wise, H.M.; Leslie, N.R. Mechanisms of PTEN loss in cancer: It’s all about diversity. Semin. Cancer Biol. 2019, 59, 66–79. [Google Scholar] [CrossRef]
- Yue, F.; Song, C.; Huang, D.; Narayanan, N.; Qiu, J.; Jia, Z.; Yuan, Z.; Oprescu, S.N.; Roseguini, B.T.; Deng, M.; et al. PTEN Inhibition Ameliorates Muscle Degeneration and Improves Muscle Function in a Mouse Model of Duchenne Muscular Dystrophy. Mol. Ther. 2021, 29, 132–148. [Google Scholar] [CrossRef]
- Kumagai, H.; Coelho, A.R.; Wan, J.; Mehta, H.H.; Yen, K.; Huang, A.; Zempo, H.; Fuku, N.; Maeda, S.; Oliveira, P.J.; et al. MOTS-c reduces myostatin and muscle atrophy signaling. Am. J. Physiol. Endocrinol. Metab. 2021, 320, E680–E690. [Google Scholar] [CrossRef]
- Park, H.; Seo, K.S.; Lee, M.; Seo, S. Identification of meat quality-related differentially methylated regions in the DNA of the longissimus dorsi muscle in pig. Anim. Biotechnol. 2020, 31, 189–194. [Google Scholar] [CrossRef]
- Zhou, W.; Li, X.; Premont, R.T. Expanding functions of GIT Arf GTPase-activating proteins, PIX Rho guanine nucleotide exchange factors and GIT-PIX complexes. J. Cell Sci. 2016, 129, 1963–1974. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Amanullah, A.; Chhangani, D.; Mishra, R.; Prasad, A.; Mishra, A. Mahogunin Ring Finger-1 (MGRN1), a Multifaceted Ubiquitin Ligase: Recent Unraveling of Neurobiological Mechanisms. Mol. Neurobiol. 2016, 53, 4484–4496. [Google Scholar] [CrossRef] [PubMed]
- Khalil, R. Ubiquitin-Proteasome Pathway and Muscle Atrophy. Adv. Exp. Med. Biol. 2018, 1088, 235–248. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, Z.; Cao, X.; He, H.; Han, S.; Chen, Y.; Cui, C.; Zhao, J.; Li, D.; Wang, Y.; et al. Circular RNA profiling identified an abundant circular RNA circTMTC1 that inhibits chicken skeletal muscle satellite cell differentiation by sponging miR-128-3p. Int. J. Biol. Sci. 2019, 15, 2265–2281. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, M.; Kong, L.; Cao, M.; Zhang, M.; Wang, Y.; Song, C.; Fang, X.; Chen, H.; Zhang, C. CircARID1A regulates mouse skeletal muscle regeneration by functioning as a sponge of miR-6368. FASEB J. 2021, 35, e21324. [Google Scholar] [CrossRef] [PubMed]



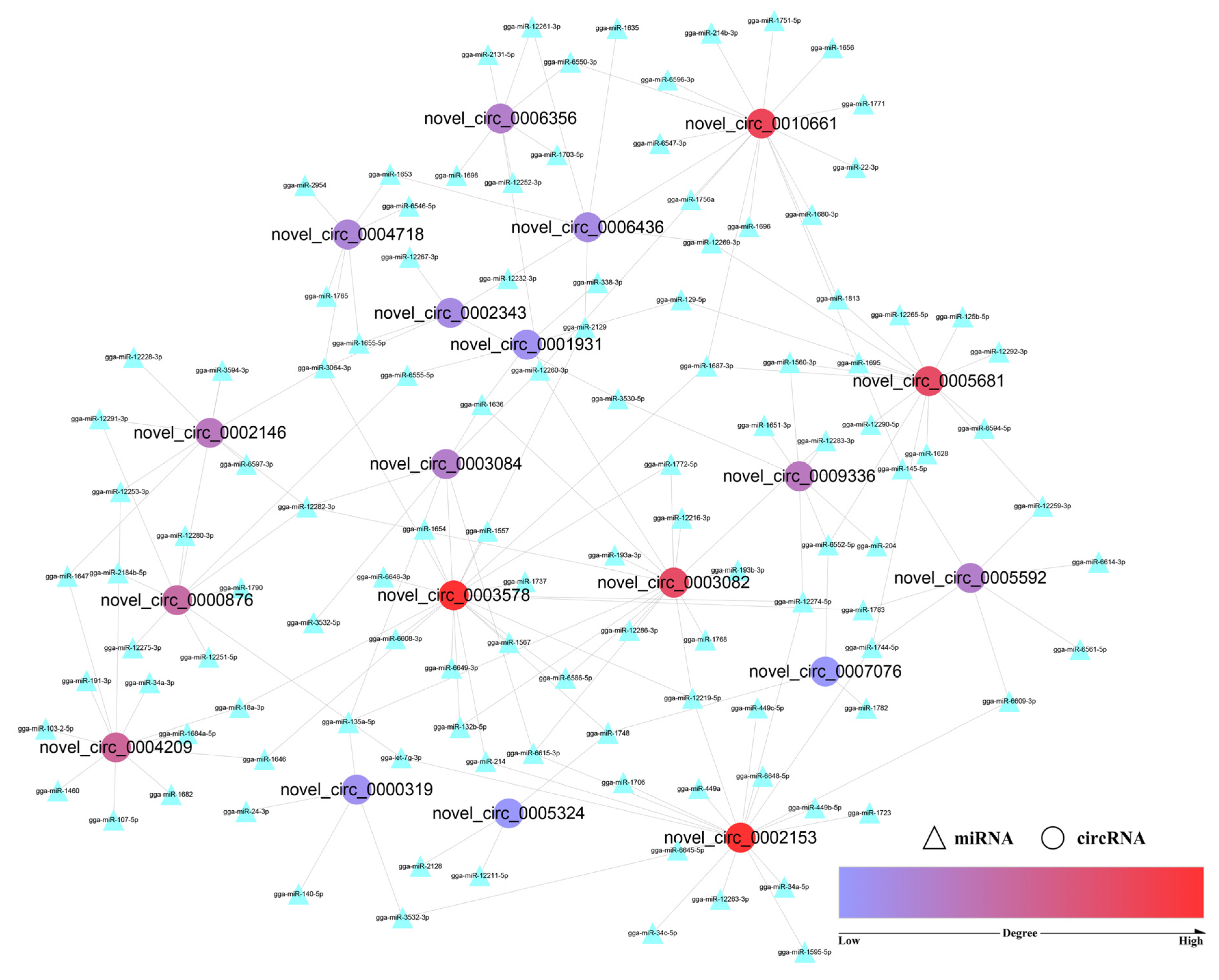
| Name | Host Gene | Degree | Name | Host Gene | Degree |
|---|---|---|---|---|---|
| novel_circ_0002153 | AHCYL2 | 19 | novel_circ_0005592 | ITSN2 | 7 |
| novel_circ_0003578 | DVL1 | 19 | novel_circ_0006356 | REV3L | 7 |
| novel_circ_0010661 | ENSGALG00000002326 | 16 | novel_circ_0004718 | SLC25A13 | 6 |
| novel_circ_0003082 | DERA | 15 | novel_circ_0002343 | DIS3 | 5 |
| novel_circ_0005681 | CDC5L | 15 | novel_circ_0006436 | RRAGD | 5 |
| novel_circ_0004209 | NCOA2 | 11 | novel_circ_0000319 | GPATCH1 | 4 |
| novel_circ_0000876 | MGRN1 | 10 | novel_circ_0001931 | CNKSR2 | 4 |
| novel_circ_0002146 | ENSGALG00000016826 | 8 | novel_circ_0005324 | MOCOS | 3 |
| novel_circ_0009336 | LRRFIP1 | 8 | novel_circ_0007076 | MOB1B | 3 |
| novel_circ_0003084 | DERA | 7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; Zhang, J.; Wu, P.; Ling, X.; Wang, Q.; Zhou, K.; Li, P.; Zhang, L.; Ye, H.; Zhang, Q.; et al. Transcriptome Sequencing Analysis of circRNA in Skeletal Muscle between Fast- and Slow-Growing Chickens at Embryonic Stages. Animals 2022, 12, 3166. https://doi.org/10.3390/ani12223166
Zhang G, Zhang J, Wu P, Ling X, Wang Q, Zhou K, Li P, Zhang L, Ye H, Zhang Q, et al. Transcriptome Sequencing Analysis of circRNA in Skeletal Muscle between Fast- and Slow-Growing Chickens at Embryonic Stages. Animals. 2022; 12(22):3166. https://doi.org/10.3390/ani12223166
Chicago/Turabian StyleZhang, Genxi, Jin Zhang, Pengfei Wu, Xuanze Ling, Qifan Wang, Kaizhi Zhou, Peifeng Li, Li Zhang, Hongxin Ye, Qi Zhang, and et al. 2022. "Transcriptome Sequencing Analysis of circRNA in Skeletal Muscle between Fast- and Slow-Growing Chickens at Embryonic Stages" Animals 12, no. 22: 3166. https://doi.org/10.3390/ani12223166
APA StyleZhang, G., Zhang, J., Wu, P., Ling, X., Wang, Q., Zhou, K., Li, P., Zhang, L., Ye, H., Zhang, Q., Wei, Q., Zhang, T., & Wang, X. (2022). Transcriptome Sequencing Analysis of circRNA in Skeletal Muscle between Fast- and Slow-Growing Chickens at Embryonic Stages. Animals, 12(22), 3166. https://doi.org/10.3390/ani12223166






