High Prevalence and Genetic Variability of Hepatozoon canis in Grey Wolf (Canis lupus L. 1758) Population in Serbia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Study Area and Sample Preparation
2.2. Sequencing and Sequence Processing
2.3. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, T.G. The genus Hepatozoon (Apicomplexa: Adeleina). J. Parasitol. 1996, 82, 565–585. Available online: https://www.jstor.org/stable/3283781 (accessed on 8 August 2021). [CrossRef] [PubMed]
- Baneth, G.; Samish, M.; Alekseev, E.; Aroch, I.; Shkap, V. Transmission of Hepatozoon canis to dogs by naturally-fed or percutaneously-injected Rhipicephalus sanguineus ticks. J. Parasitol. 2001, 87, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Hodžić, A.; Georges, I.; Postl, M.; Duscher, G.G.; Jeschke, D.; Szentiks, A.C.; Ansorge, H.; Heddergott, M. Molecular survey of tick-borne pathogens reveals a high prevalence and low genetic variability of Hepatozoon canis in free-ranging grey wolves (Canis lupus) in Germany. Ticks Tick-Borne Dis. 2020, 11, 101389. [Google Scholar] [CrossRef] [PubMed]
- Mitková, B.; Hrazdilová, K.; D’Amico, G.; Duscher, G.G.; Suchentrunk, F.; Forejtek, P.; Gherman, C.M.; Matei, I.A.; Ionică, A.M.; Daskalaki, A.A.; et al. Eurasian golden jackal as host of canine vector-borne protists. Parasit. Vectors 2017, 10, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penzhorn, B.L.; Netherlands, E.C.; Cook, C.A.; Smit, N.J.; Vorster, I.; Harrison-White, R.F.; Oosthuizen, M.C. Occurrence of Hepatozoon canis (Adeleorina: Hepatozoidae) and Anaplasma spp. (Rickettsiales: Anaplasmataceae) in black-backed jackals (Canis mesomelas) in South Africa. Parasit. Vectors 2018, 1, 158. [Google Scholar] [CrossRef] [Green Version]
- André, M.R.; Adania, C.H.; Teixeira, R.H.F.; Vargas, G.H.; Falcade, M.; Sousa, L.; Salles, A.R.; Allegretti, S.M.; Filippe, P.A.N.; Machado, R.Z. Molecular detection of Hepatozoon spp. In Brazilian and exotic wild carnivores. Vet. Parasitol. 2010, 173, 134–138. [Google Scholar] [CrossRef] [PubMed]
- Criado-Fornelio, A.; Martinez-Marcos, A.; Buling-Sarana, A.; Barba-Carretero, J.C. Molecular studies on Babesia, Theileria and Hepatozoon in southern Europe. Part I. Epizootiological aspects. Vet. Parasitol. 2003, 113, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, L.; Cortes, H.C.E.; Eyal, O.; Reis, A.; Lopes, A.P.; Vila-Viçosa, M.J.; Rodrigues, P.A.; Baneth, G. Molecular and histopathological detection of Hepatozoon canis in red foxes (Vulpes vulpes) from Portugal. Parasit. Vectors 2014, 7, 113. Available online: http://www.parasitesandvectors.com/content/7/1/113 (accessed on 8 August 2021). [CrossRef] [PubMed] [Green Version]
- Baneth, G.; Barta, J.R.; Martin, D.S.; Macintire, D.K.; Vincent-Johnson, N. Genetic and antigenic evidence supports the separation of Hepatozoon canis and Hepatozoon americanum at the species level. J. Clin. Microbiol. 2000, 38, 1298–1301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baneth, G.; Harmelin, A.; Presentey, B. Hepatozoon canis infection in two dogs. J. Am. Vet. Med. Assoc. 1995, 206, 1891–1894. [Google Scholar] [PubMed]
- Baneth, G.; Weigler, B. Retrospective casecontrol study of hepatozoonosis in dogs in Israel. J. Vet. Intern. Med. 1997, 11, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Otranto, D.; Dantas-Torres, F.; Weigl, S.; Latrofa, M.; Stanneck, D.; Decaprariis, D.; Capelli, G.; Baneth, G. Diagnosis of Hepatozoon canis in young dogs by cytology and PCR. Parasit. Vectors 2011, 4, 55. Available online: http://www.parasitesandvectors.com/content/4/1/55 (accessed on 8 August 2021). [CrossRef] [PubMed] [Green Version]
- Baneth, G.; Mathew, J.S.; Shkap, V.; Macintire, D.K.; Barta, J.R.; Ewing, S.A. Canine hepatozoonosis—Two disease syndromes caused by separate Hepatozoon species. Trends Parasitol. 2003, 19, 27–31. [Google Scholar] [CrossRef] [PubMed]
- Baneth, G. Perspectives on canine and feline hepatozoonosis. Vet. Parasitol. 2011, 181, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Inoue, M.; Tateyama, S.; Taura, Y.; Nakama, S. Vertical transmission of Hepatozoon canis in dogs. J. Vet. Med. Sci. 1993, 55, 867–868. [Google Scholar] [CrossRef] [Green Version]
- Baneth, G.; Samish, M.; Shkap, V. Life cycle of Hepatozoon canis (Apicomplexa: Adeleorina: Hepatozoidae) in the tick Rhipicephalus sanguineus and domestic dog (Canis familiaris). J. Parasitol. 2007, 93, 283–299. [Google Scholar] [CrossRef] [PubMed]
- Juwaid, S.; Sukara, R.; Penezić, A.; Mihaljica, D.; Veinović, G.; Kavallieratos, G.N.; Ćirović, D.; Tomanović, S. First Evidence of Tick-Borne Protozoan Pathogens, Babesia sp. and Hepatozoon canis, In Red Foxes (Vulpes Vulpes) In Serbia. Acta Vet. Hung. 2019, 67, 70–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boitani, L. Action Plan for the Conservation of the Wolves (Canis lupus) in Europe; Convention on the Conservation of European Wildlife and Natural Habitats (Bern Convention); Nature and Environment, Council of Europe Publishing: Strasbourg, France, 2000; pp. 7–84. [Google Scholar]
- Cahapron, G.; Kaczensky, P.; Linnell, J.D.C.; von Arx, M.; Huber, D.J.; Andrén, H.; López-Bao, J.V.; Adamec, A.; Álvares, F.; Anders, O.; et al. Recovery of large carnivores in Europe’s modern human-dominated landscapes. Science 2014, 346, 1517–1519. [Google Scholar] [CrossRef] [Green Version]
- Cimatti, M.; Ranc, N.; Benítez-López, A.; Maiorano, L.; Boitani, L.; Cagnacci, F.; Čengić, M.; Ciucci, P.; Huijbregts, M.A.J.; Krofel, M.; et al. Large carnivore expansion in Europe is associated with human population density and land cover changes. Divers. Distrib. 2021, 27, 602–617. [Google Scholar] [CrossRef]
- Reinhardt, I.; Kluth, G.; Nowak, S.; Myslajek, R. Standards for the monitoring of the Central European wolf population in Germany and Poland; German Federal Agency for Nature Conservation: Bonn, Germany, 2015; Volume 398, p. 43. [Google Scholar]
- Inokuma, H.; Okuda, M.; Ohno, K.; Shimoda, K.; Onishi, T. Analysis of the 18S rRNA gene sequence of a Hepatozoon detected in two Japanese dogs. Vet. Parasitol. 2002, 106, 265–271. [Google Scholar] [CrossRef]
- National Center for Biotechnology. Available online: http://www.ncbi.nlm.nih.gov/BLAST (accessed on 27 July 2021).
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Mega, X. Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hodžić, A.; Alić, A.; Fuehrer, H.P.; Harl, J.; Wille-Piazzai, W.; Duscher, G.G. A molecular survey of vector-borne pathogens in red foxes (Vulpes vulpes) from Bosnia and Herzegovina. Parasit. Vectors 2015, 8, 88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farkas, R.; Solymosi, N.; Takács, N.; Hornyák, A.; Hornok, S.; Nachum-Biala, Y.; Baneth, G. First molecular evidence of Hepatozoon canis infection in red foxes and golden jackals from Hungary. Parasit. Vectors 2014, 7, 303. [Google Scholar] [CrossRef] [Green Version]
- Helm, S.C.; Samson-Himmelstjerna, V.G.; Liesner, M.J.; Kohn, B.; Müller, E.; Schaper, R.; Pachnicke, S.; Schulze, C.; Krücken, J. Identical 18S rRNA haplotypes of Hepatozoon canis in dogs and foxes in Brandenburg, Germany. Ticks Tick-Borne Dis. 2020, 11, 101520. [Google Scholar] [CrossRef]
- Hodžić, A.; Alić, A.; Prašović, S.; Otranto, D.; Baneth, G.; Duscher, G.G. Hepatozoon silvestris sp. nov.: Morphological and molecular characterization of a new species of Hepatozoon (Adeleorina: Hepatozoidae) from the European wild cat (Felis silvestris silvestris). Parasitology 2017, 144, 650–661. [Google Scholar] [CrossRef] [Green Version]
- Hornok, S.; Sándor, D.A.; Tomanović, S.; Beck, R.; D’Amico, G.; Kontschán, J.; Takács, N.; Görföl, T.; Bendjeddou, L.M.; Földvári, G.; et al. East and west separation of Rhipicephalus sanguineus mitochondrial lineages in the Mediterranean Basin. Parasit. Vectors 2017, 10, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemtsova, E.G.; Apanaskevich, A.D.; Reeves, K.W.; Hahn, M.; Snellgrove, A.; Levin, L.M. Phylogeography of Rhipicephalus sanguineus sensu lato and its relationships with climatic factors. Exp. Appl. Acarol. 2016, 69, 191–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloboda, M.; Kamler, M.; Bulantová, J.; Votýpka, J.; Modrý, D. Rodents as intermediate hosts of Hepatozoon ayorgbor (Apicomplexa: Adeleina: Hepatozoidae) from the African ball python, Python regius? Folia Parasitol. 2008, 55, 13–16. [Google Scholar] [CrossRef] [Green Version]
- Ćirović, D.; Penezić, A. Importance of slaughter waste in winter diet of wolves (Canis lupus) in Serbia. North. West. J. Zool. 2019, 15, 175–178. [Google Scholar]
- Castillo-Contreras, R.; Magen, L.; Birtles, R.; Varela-Castro, L.; Hall, J.L.; Conejero, C.; Aguilar, X.F.; Colom-Cadena, A.; Lavín, S.; Mentaberre, G.; et al. Ticks on wild boar in the metropolitan area of Barcelona (Spain) are infected with spotted fever group rickettsiae. Transbound. Emerg. Dis. 2021, 69, e82–e95. [Google Scholar] [CrossRef]
- Accorsi, A.; Schiavetti, I.; Listorti, V.; Dellepiane, M.; Masotti, C.; Ercolini, C.; Guardone, L.; Razzuoli, E. Hard Ticks (Ixodidae) from Wildlife in Liguria, Northwest Italy: Tick Species Diversity and Tick-Host Associations. Insects 2022, 13, 199. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, G.; Girardi, M.; Cagnacci, F.; Devineau, O.; Tagliapietra, V. First Record of Hepatozoon spp. in Alpine Wild Rodents: Implications and Perspectives for Transmission Dynamics across the Food Web. Microorganisms 2022, 10, 712. [Google Scholar] [CrossRef] [PubMed]
- Rund, D.; Neves, V.; Quillfeldt, P. Molecular survey of Hepatozoon infection of Teira dugesii in the Azores. Anim. Biodivers. Conserv. 2019, 42, 19–29. [Google Scholar] [CrossRef]
 ) and negative (
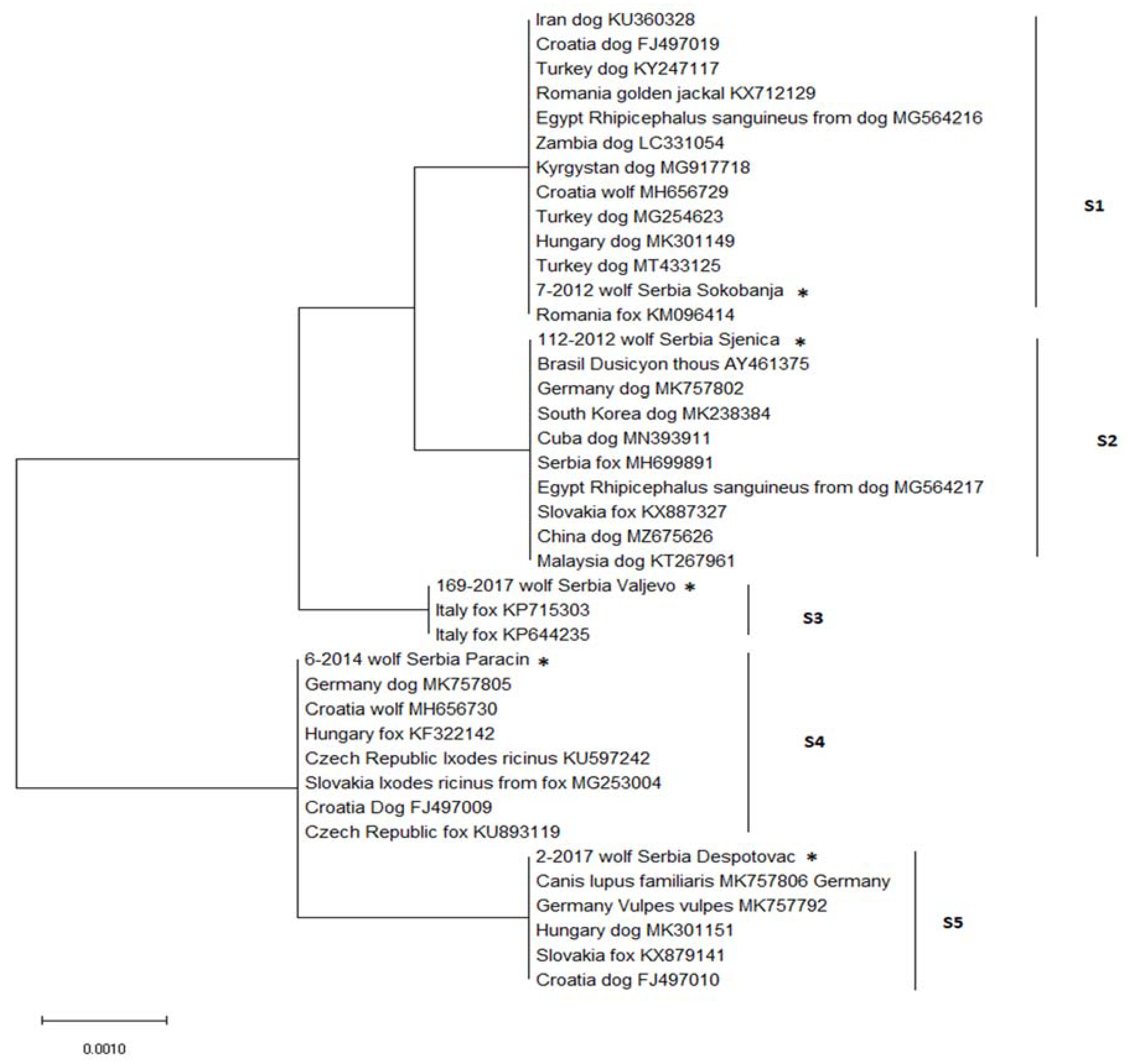
) and negative ( ), indicate the finding of DNA of Hepatozoon canis in spleen samples from each locality. Charts present distribution of sequence types S1–S5 in western (left) and eastern (right) part of Serbia).
), indicate the finding of DNA of Hepatozoon canis in spleen samples from each locality. Charts present distribution of sequence types S1–S5 in western (left) and eastern (right) part of Serbia).
 ) and negative (
) and negative ( ), indicate the finding of DNA of Hepatozoon canis in spleen samples from each locality. Charts present distribution of sequence types S1–S5 in western (left) and eastern (right) part of Serbia).
), indicate the finding of DNA of Hepatozoon canis in spleen samples from each locality. Charts present distribution of sequence types S1–S5 in western (left) and eastern (right) part of Serbia).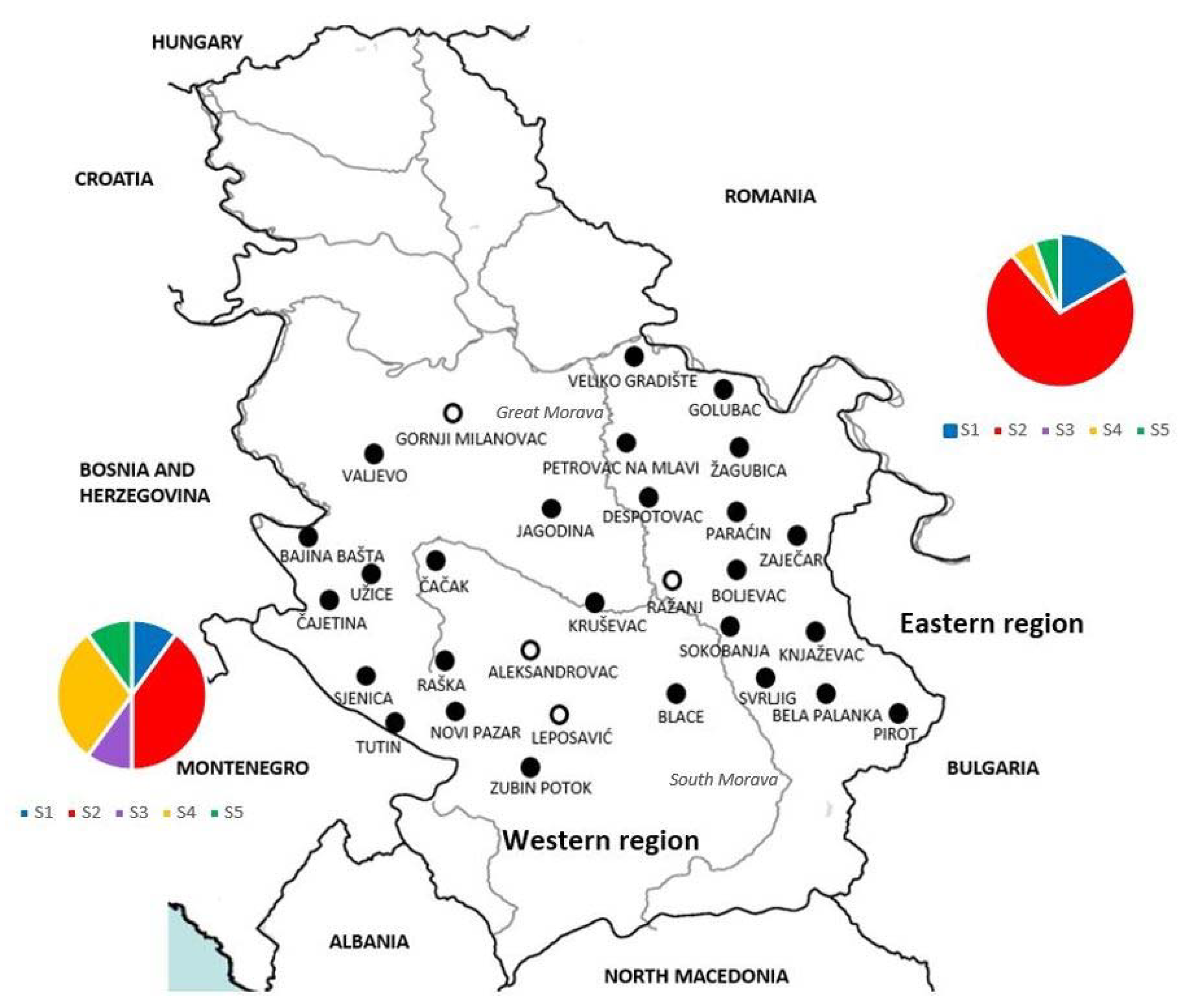
| Region | Locality | No. of Collected Samples | M/F | No. of Positive Samples Hepatozoon canis (%) | M/F |
|---|---|---|---|---|---|
| WEST REGION | Bajina Bašta | 2 | 2/0 | 1 (50%) | 1/0 |
| Valjevo | 2 | 2/0 | 2 (100%) | 2/0 | |
| Gornji Milanovac | 1 | 1/0 | / | 0/0 | |
| Čajetina | 7 | 5/2 | 4 (57.14%) | 3/1 | |
| Užice | 6 | 5/1 | 3 (50%) | 3/0 | |
| Čačak | 2 | 1/1 | 1 (50%) | 1/0 | |
| Sjenica | 30 | 17/13 | 11 (36.66%) | 6/5 | |
| Raška | 1 | 0/1 | 1 (100%) | 0/1 | |
| Aleksandrovac | 1 | 1/0 | / | 0/0 | |
| Kruševac | 2 | 2/0 | 2 (100%) | 2/0 | |
| Novi Pazar | 3 | 2/1 | 1 (33,.33%) | 1/0 | |
| Leposavić | 1 | 0/1 | / | 0/0 | |
| Blace | 1 | 1/0 | 1 (100%) | 1/0 | |
| Zubin Potok | 1 | 1//0 | 1 (100%) | 1/0 | |
| Tutin | 2 | 2/0 | 1 (50%) | 1/0 | |
| Jagodina | 1 | 1/0 | 1 (100%) | 1/0 | |
| Total | 63 | 30 (47.6%) | |||
| EAST REGION | Veliko Gradište | 2 | 1/1 | 1 (50%) | 1/0 |
| Golubac | 3 | 1/2 | 3 (100%) | 1/2 | |
| Petrovac na Mlavi | 5 | 3/2 | 5 (100%) | 3/2 | |
| Žagubica | 7 | 4/3 | 2 (28.57%) | 1/1 | |
| Despotovac | 4 | 0/4 | 3 (75%) | 0/3 | |
| Paraćin | 6 | 1/5 | 6 (100%) | 1/5 | |
| Zaječar | 2 | 1/1 | 1 (50%) | 0/1 | |
| Boljevac | 1 | 1/0 | 1 (100%) | 1/0 | |
| Ražanj | 2 | 1/1 | / | 0/0 | |
| Sokobanja | 1 | 1/0 | 1 (100%) | 1/0 | |
| Knjaževac | 4 | 3/1 | 3 (75%) | 3/0 | |
| Svrljig | 2 | 1/1 | 2 (100%) | 1/1 | |
| Pirot | 3 | 2/1 | 3 (100%) | 2/1 | |
| Bela Palanka | 2 | 0/2 | 1 (50%) | 0/1 | |
| Total | 44 | 32 (72.7%) | |||
| Σ | 107 | 62 (57.94%) |
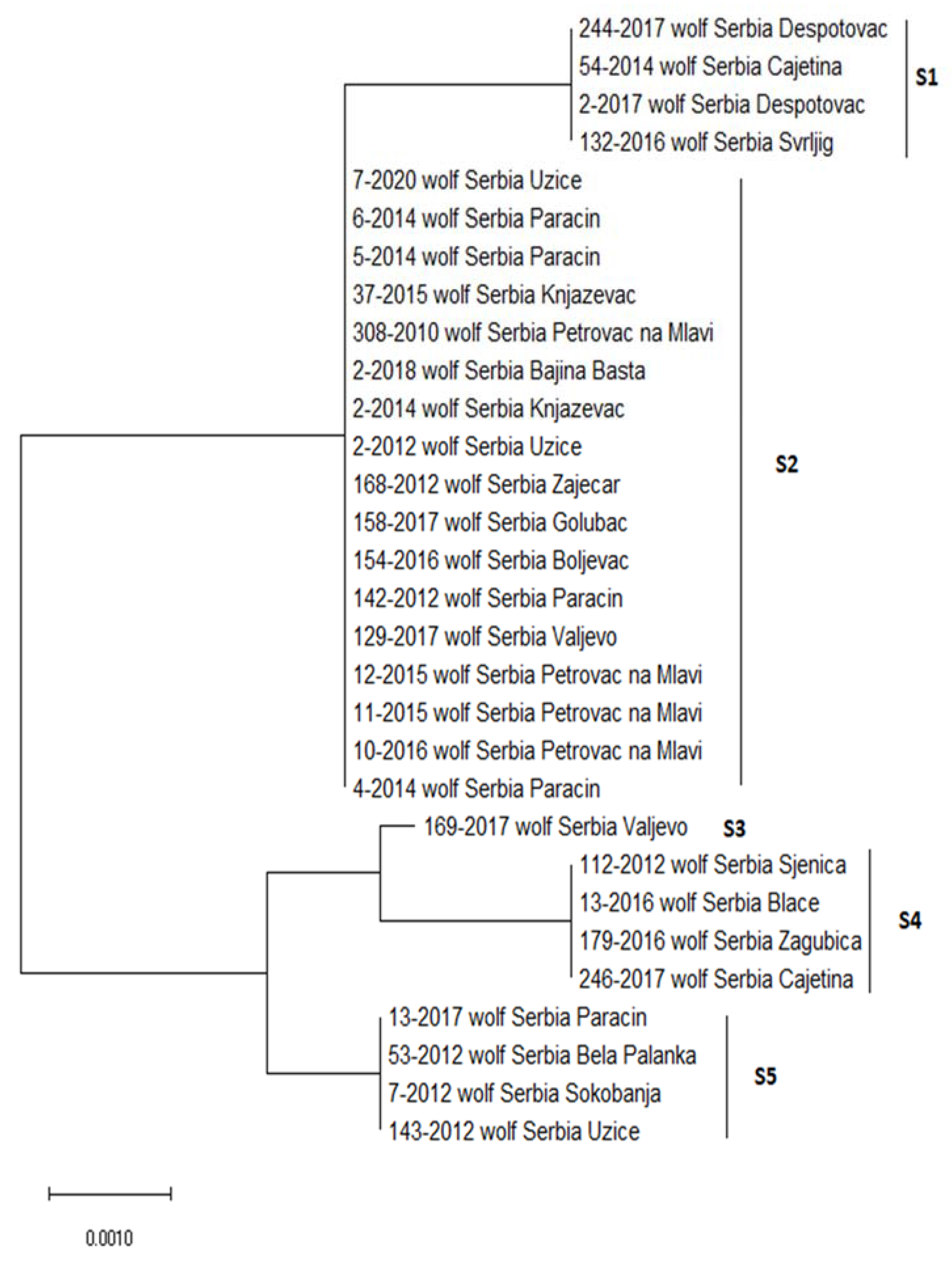
| Sequence Type | Position of the Variable Site | ||||
|---|---|---|---|---|---|
| 3 bp | 380 bp | 400 bp | 522 bp | 523 bp | |
| consensus | T | A | G | C | A |
| S1 | |||||
| S2 | A | ||||
| S3 | A | G | T | G | |
| S4 | A | G | T | T | G |
| S5 | A | G | T | T | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuručki, M.; Tomanović, S.; Sukara, R.; Ćirović, D. High Prevalence and Genetic Variability of Hepatozoon canis in Grey Wolf (Canis lupus L. 1758) Population in Serbia. Animals 2022, 12, 3335. https://doi.org/10.3390/ani12233335
Kuručki M, Tomanović S, Sukara R, Ćirović D. High Prevalence and Genetic Variability of Hepatozoon canis in Grey Wolf (Canis lupus L. 1758) Population in Serbia. Animals. 2022; 12(23):3335. https://doi.org/10.3390/ani12233335
Chicago/Turabian StyleKuručki, Milica, Snežana Tomanović, Ratko Sukara, and Duško Ćirović. 2022. "High Prevalence and Genetic Variability of Hepatozoon canis in Grey Wolf (Canis lupus L. 1758) Population in Serbia" Animals 12, no. 23: 3335. https://doi.org/10.3390/ani12233335
APA StyleKuručki, M., Tomanović, S., Sukara, R., & Ćirović, D. (2022). High Prevalence and Genetic Variability of Hepatozoon canis in Grey Wolf (Canis lupus L. 1758) Population in Serbia. Animals, 12(23), 3335. https://doi.org/10.3390/ani12233335






