Integration of mRNA and miRNA Profiling Reveals Heterosis in Oreochromis niloticus × O. aureus Hybrid Tilapia
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
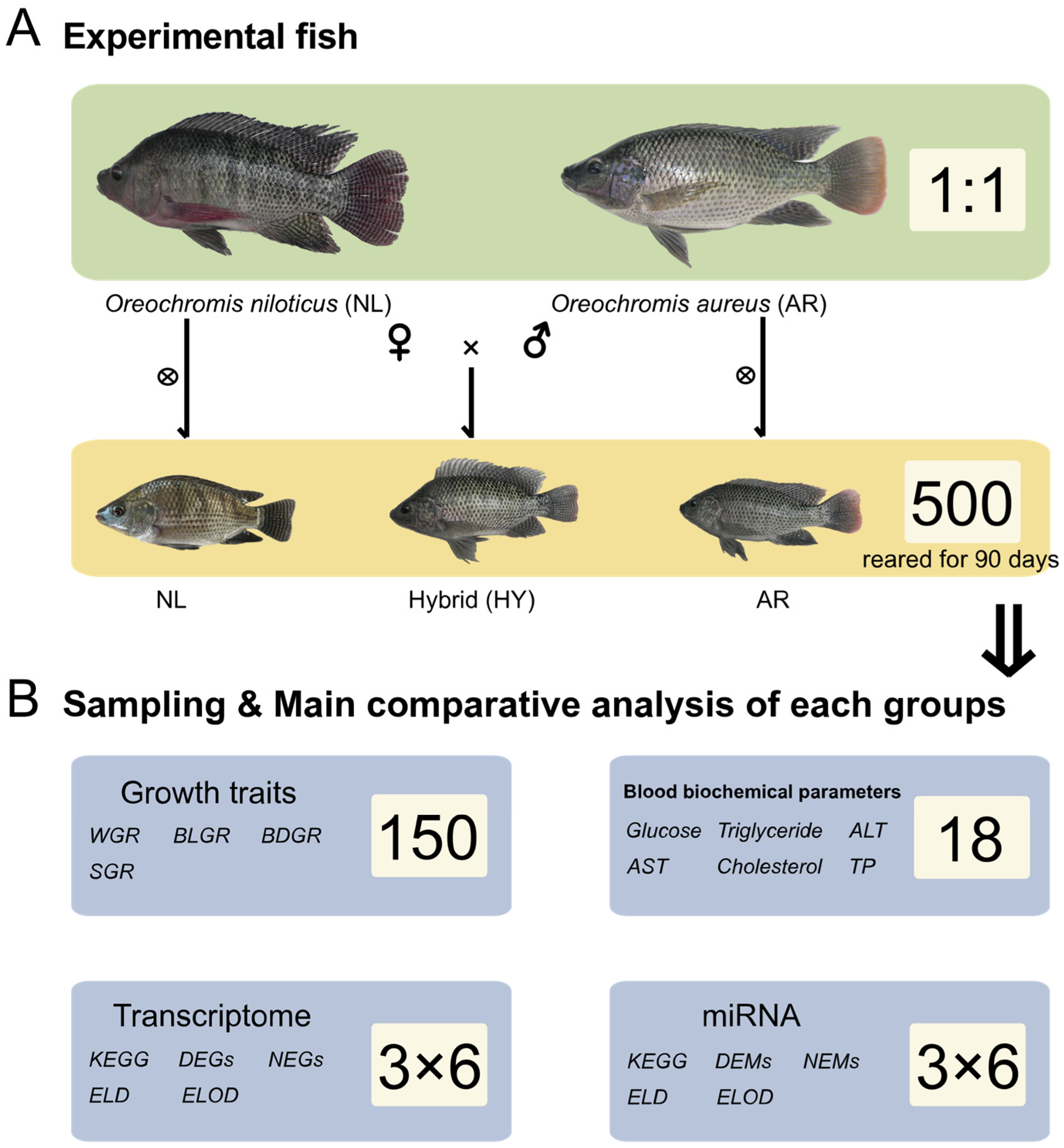
2.2. Experimental Fish Artificial Breeding and Culture
2.3. Statistical Analyses and Sample Collection
2.4. Total RNA Extraction and Quantitation
2.5. cDNA Library Construction and mRNA Sequencing
2.6. Transcriptome Sequence Alignment, Assembly, and Functional Annotation
2.7. Small RNA Library Construction and miRNA Sequencing
2.8. Identification of miRNAs
2.9. Differential Expression Analysis and mRNAs and miRNAs
2.10. MiRNA Target Gene Prediction and miRNA–mRNA Interaction
3. Results
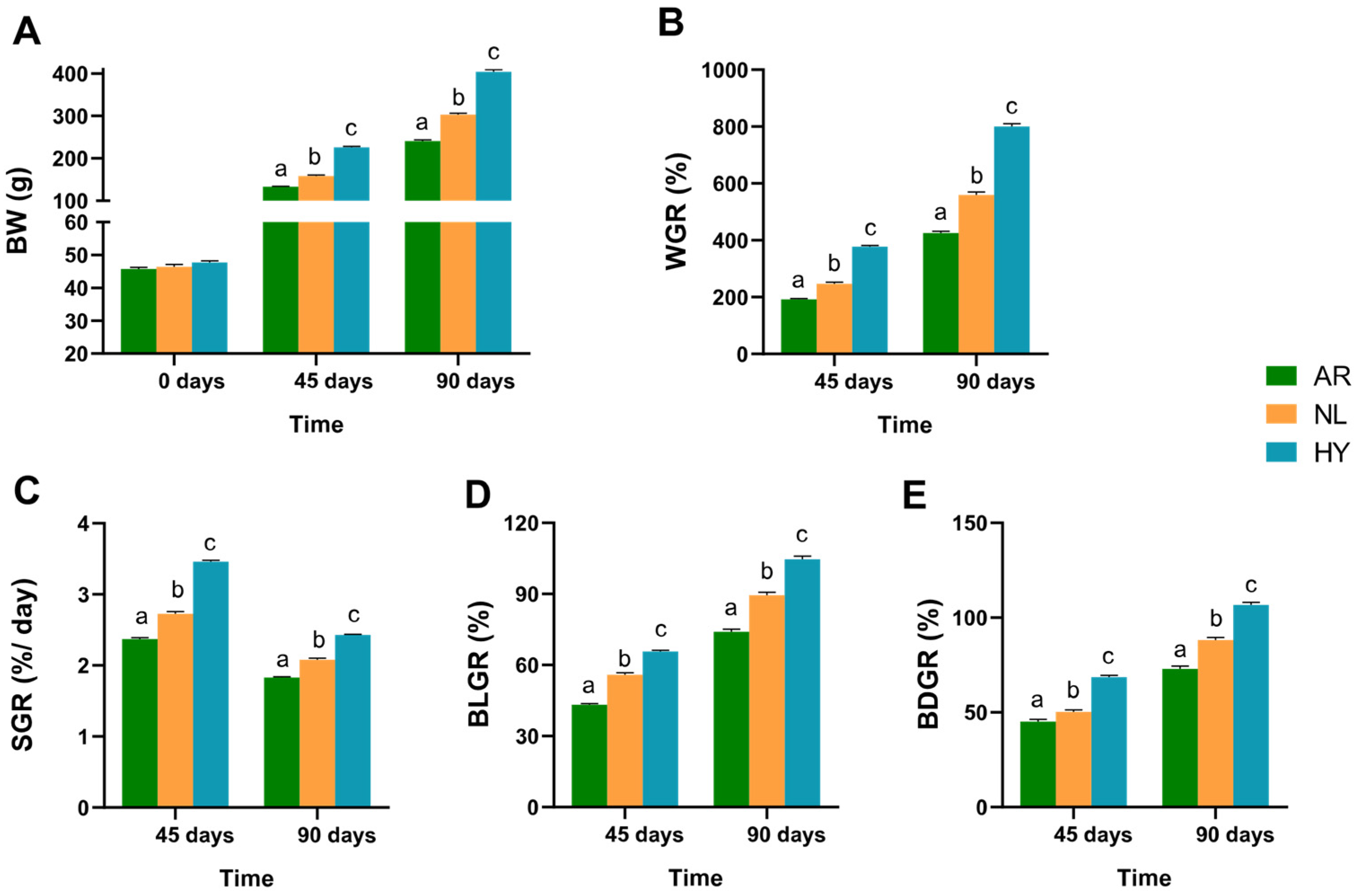
3.1. Comparison of Growth Characteristics
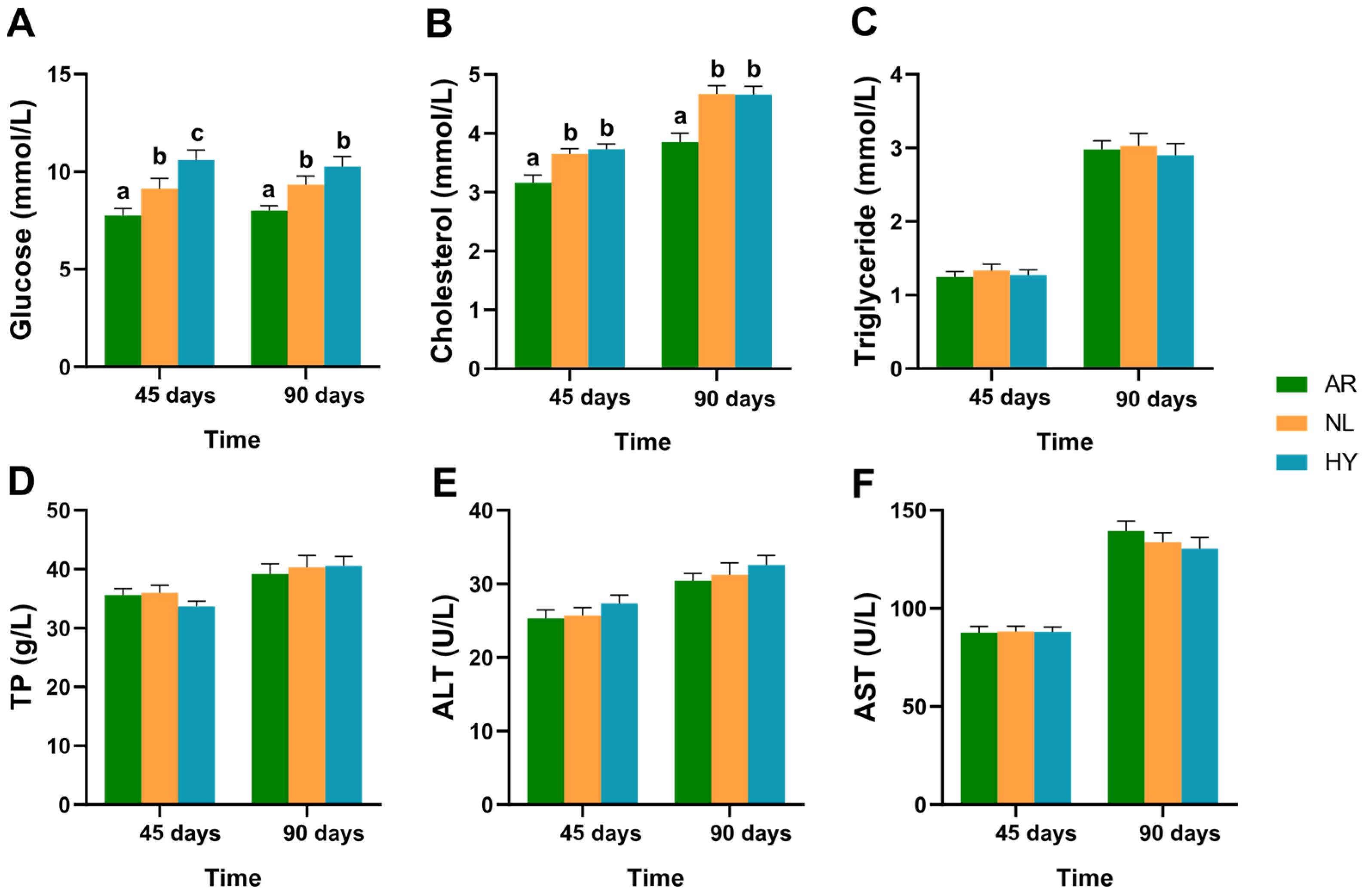
3.2. Hematological Parameter Variation
3.3. Summary of Transcriptomes and miRNAomes
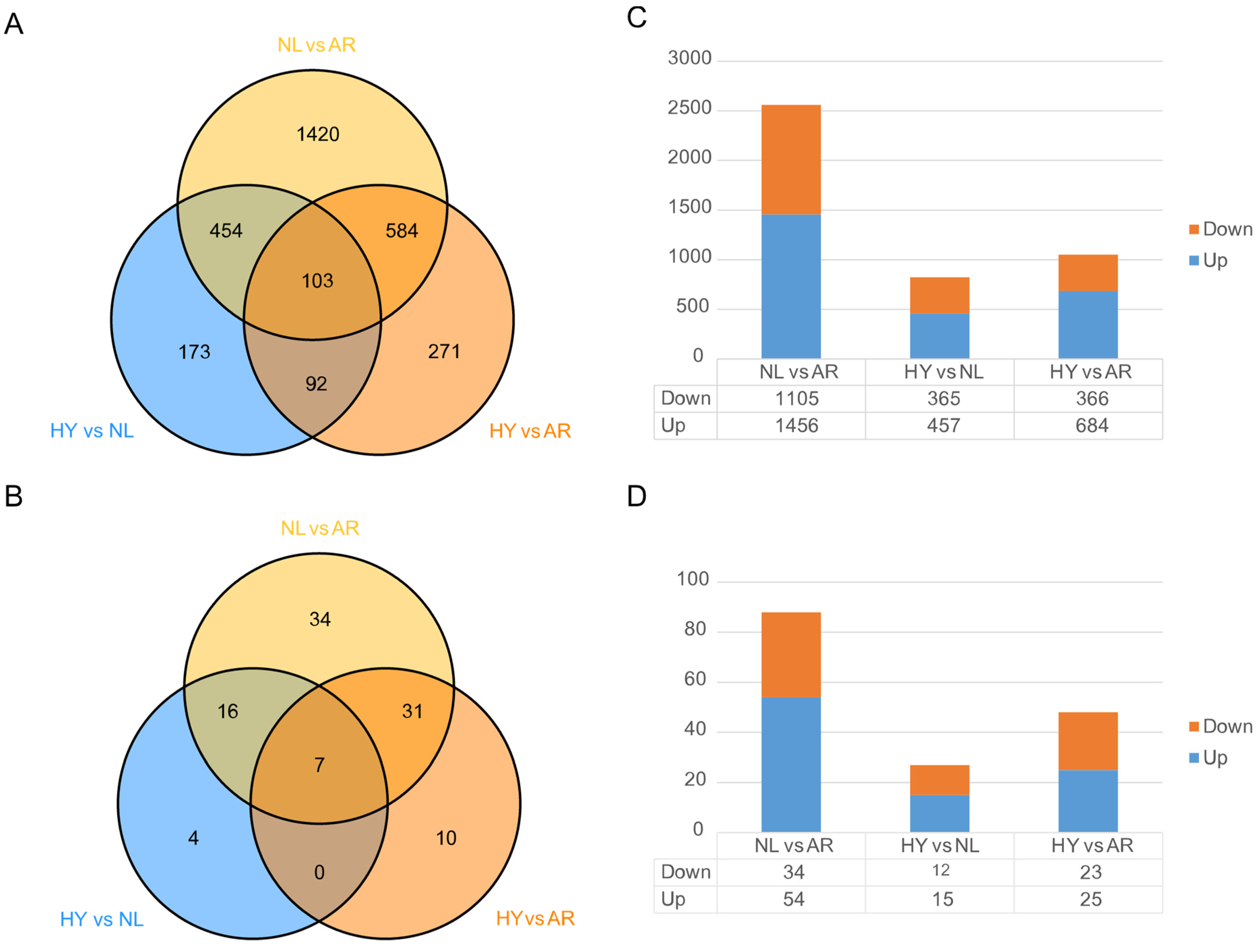
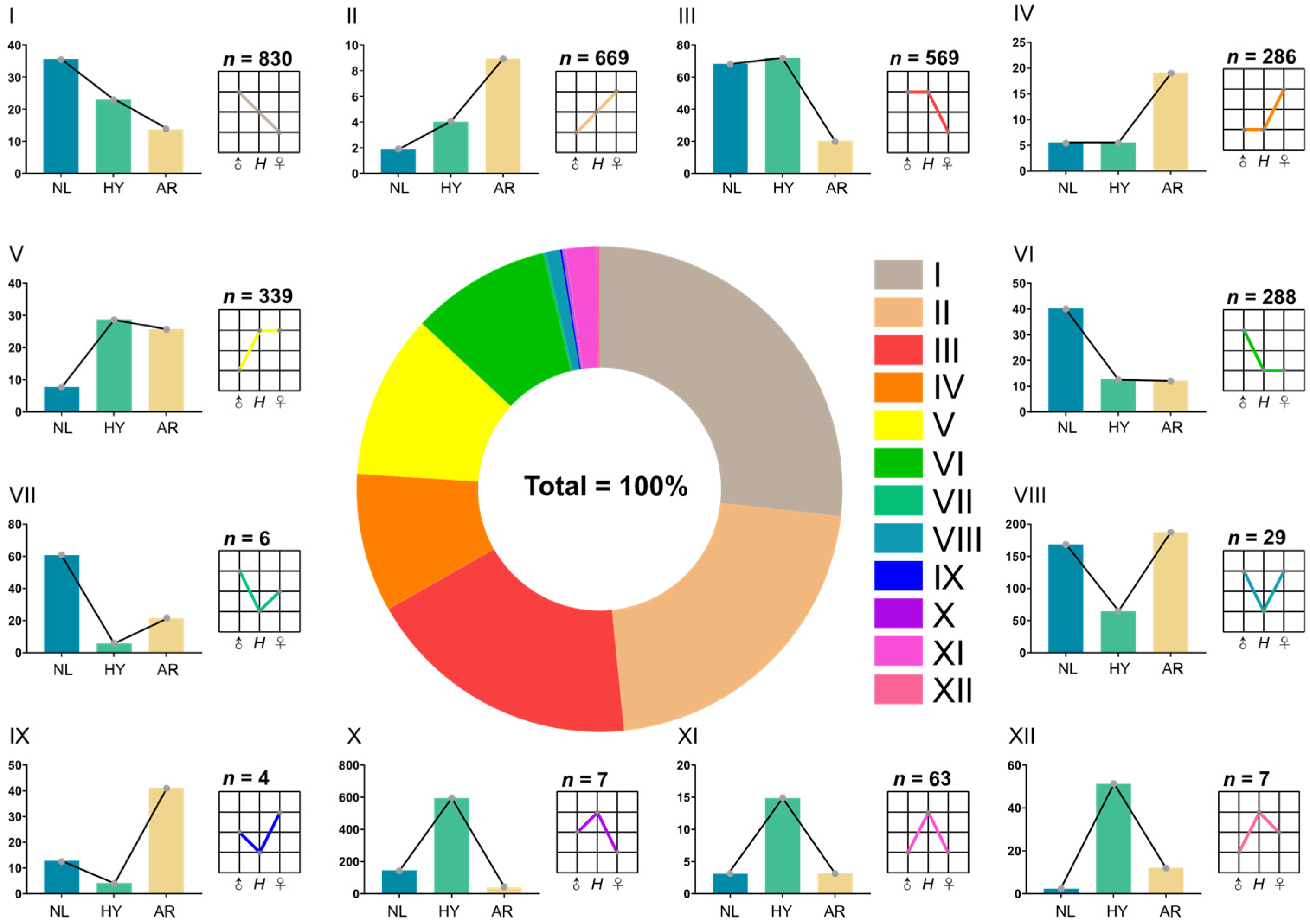
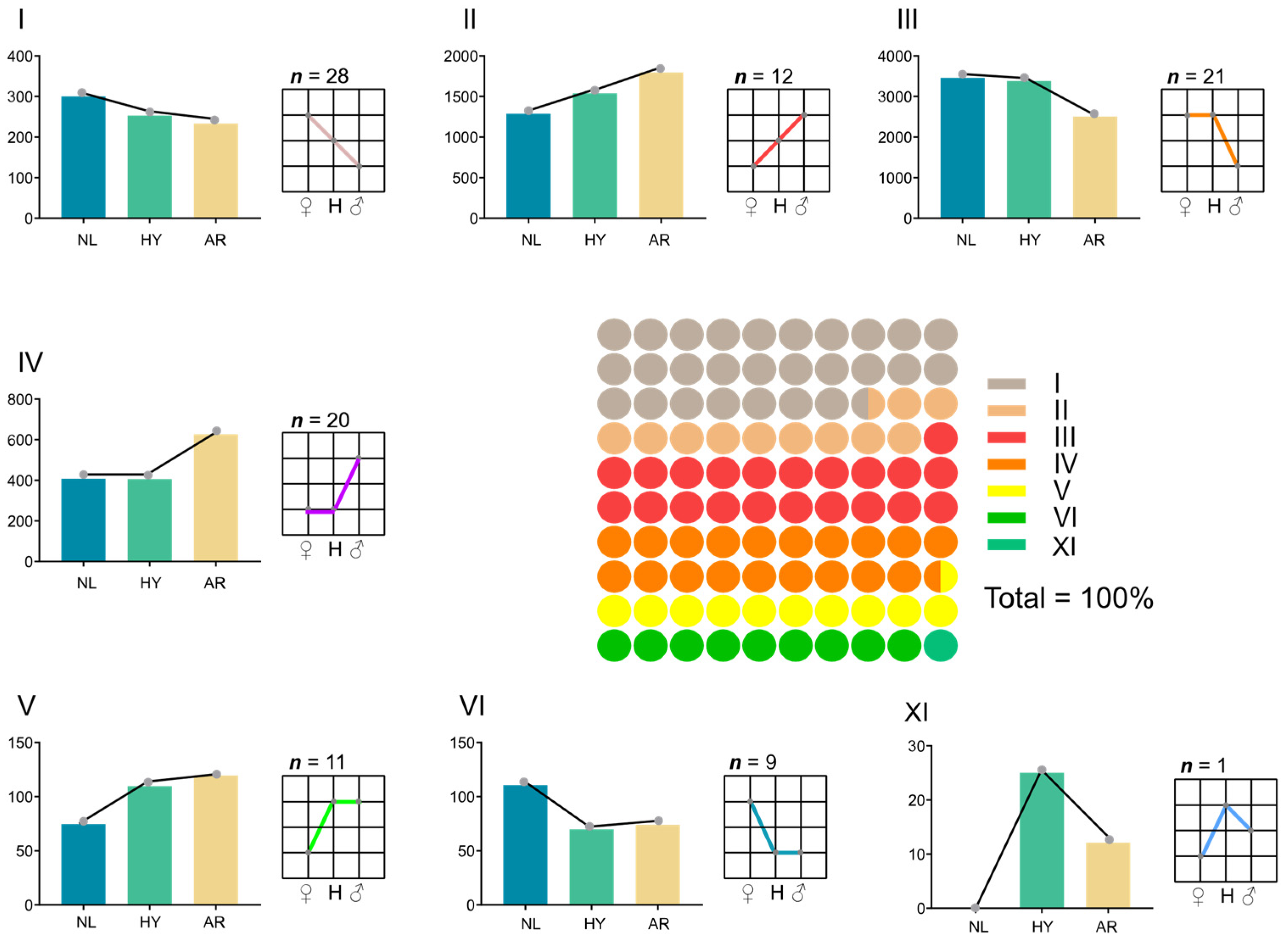
3.4. Statistics of Differentially Expressed Genes (DEGs) and miRNAs (DEMs)
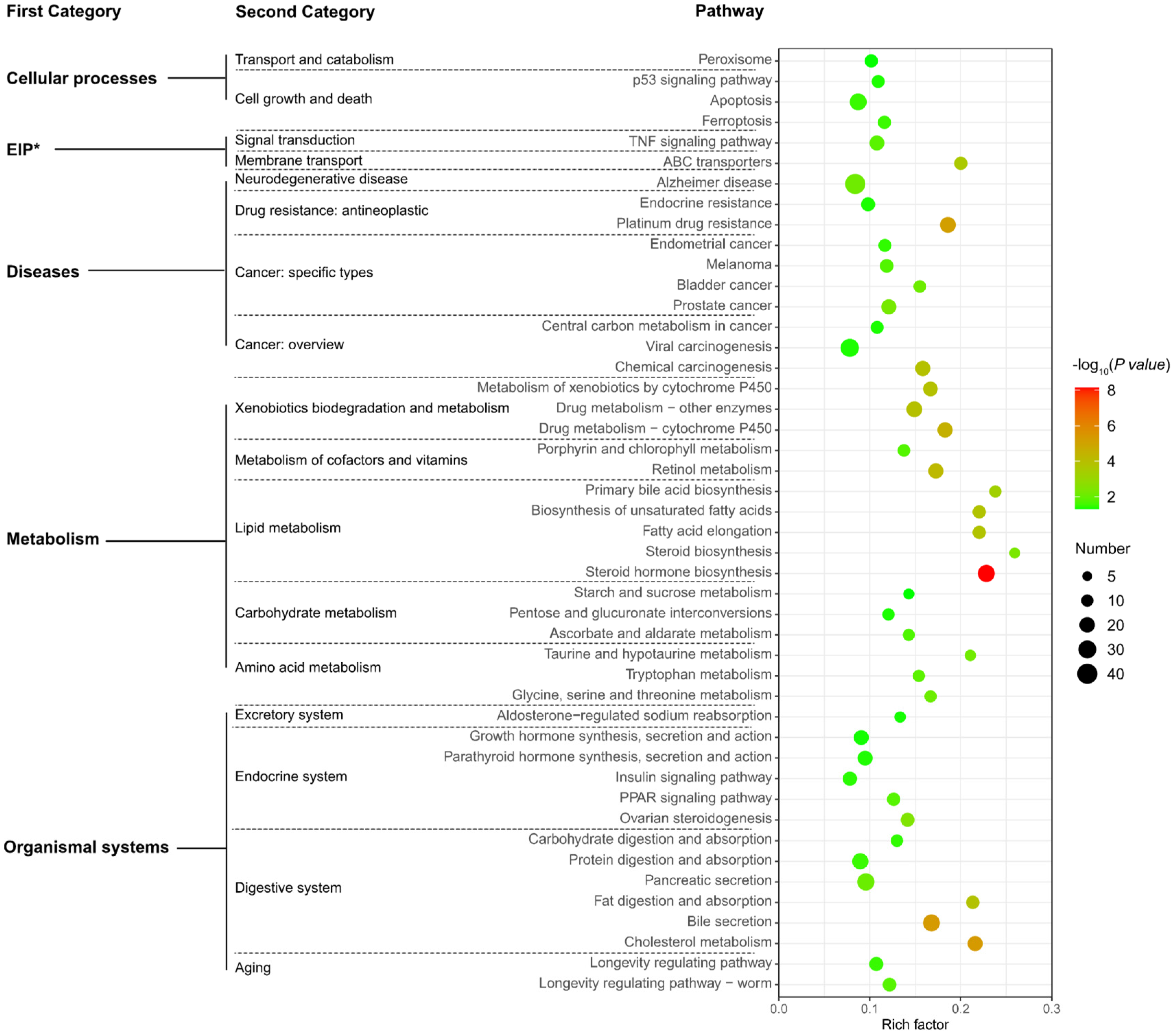
3.5. Functional Analysis of NEGs
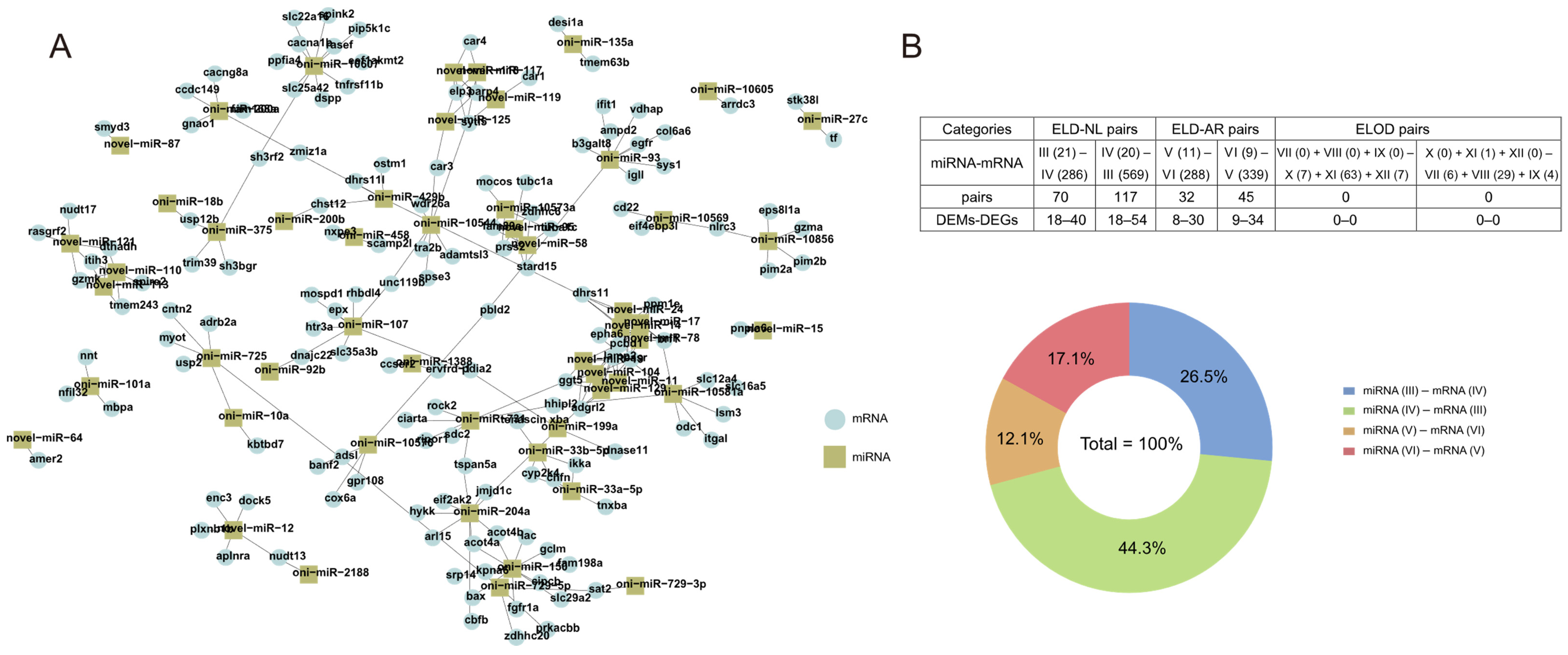
3.6. Interactions between DEMs and DEGs
4. Discussions
4.1. Improved Performance in Growth and Hematological Metabolic Indices in Hybrid Tilapia
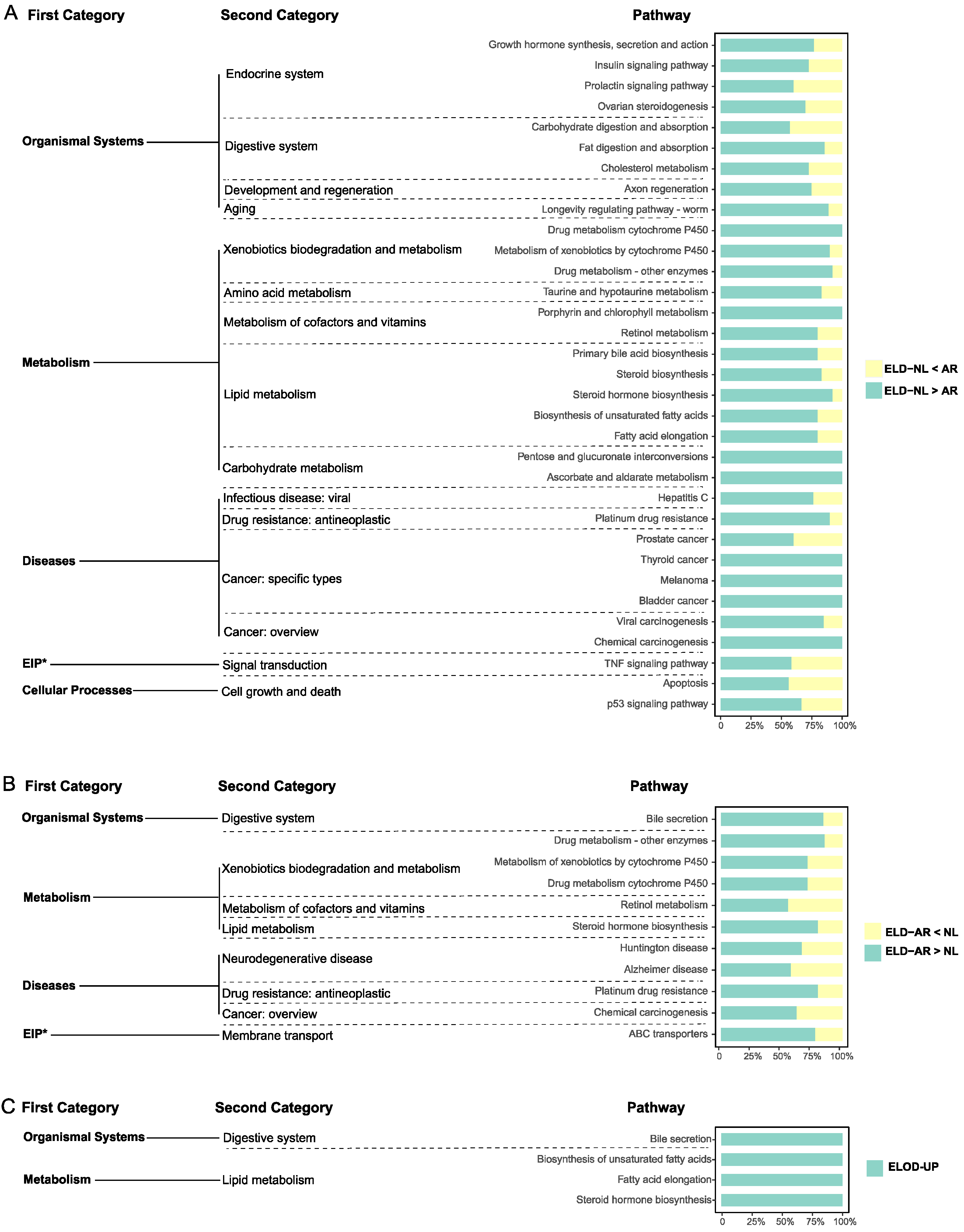
4.2. Roles of NEGs and NEMs in Formation of Super-Parental Vigor in Hybrid Tilapia
4.3. The Significance of Gene ELD and ELOD Patterns
4.3.1. Growth and Development
4.3.2. Metabolism
4.3.3. Immunity and Antioxidant Capacity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhang, F.; Gao, F.; Zeng, L.; Lu, X.; Zhao, X.; Lv, J.; Su, X.; Liu, L.; Dai, M. Transcriptome and genome sequencing elucidates the molecular basis for the high yield and good quality of the hybrid rice variety Chuanyou6203. Sci. Rep. 2020, 10, 19935. [Google Scholar] [CrossRef] [PubMed]
- Quénon, J.; Magne, M. Milk, fertility and udder health performance of purebred Holstein and three-breed rotational crossbred cows within French farms: Insights on the benefits of functional diversity. Animals 2021, 11, 3414. [Google Scholar] [CrossRef] [PubMed]
- Hubbs, C.L. Hybridization between fish species in nature. Syst. Zool. 1955, 4, 1–20. [Google Scholar] [CrossRef]
- Liu, S.; Luo, J.; Chai, J.; Ren, L.; Zhou, Y.; Huang, F.; Liu, X.; Chen, Y.; Zhang, C.; Tao, M.; et al. Genomic incompatibilities in the diploid and tetraploid offspring of the goldfish × common carp cross. Proc. Natl. Acad. Sci. USA 2016, 113, 1327–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Šimková, A.; Janáč, M.; Hyršl, P.; Krasnovyd, V.; Vetešník, L. Vigour-related traits and immunity in hybrids of evolutionary divergent cyprinoid species: Advantages of hybrid heterosis? J. Fish Biol. 2021, 98, 1155–1171. [Google Scholar] [CrossRef]
- Zhang, Y.; Pan, X.; Yin, Y.; Yang, J.; Wang, X. Fertilization and growth performance in reciprocal hybrids of Dianchi golden-line barbel (Sinocyclocheilus grahami) and domestic common carp (Cyprinus carpio) and crucian carp (Carassius auratus). Aqua Rep. 2021, 21, 100893. [Google Scholar] [CrossRef]
- Easter, E.M. Heterosis. Genetics 1936, 21, 375–397. [Google Scholar] [CrossRef]
- Jones, D.F. Dominance of linked factors as a means of accounting for heterosis. Proc. Natl. Acad. Sci. USA 1917, 3, 310–312. [Google Scholar] [CrossRef] [Green Version]
- Wright, S.; Maxson, L. Evolution and the genetics of populations. Physiol. Biochem. Zool. 1968, 8, 1191–1192. [Google Scholar] [CrossRef]
- Shen, Y.; Sun, S.; Hua, S.; Shen, E.; Ye, C.; Cai, D.; Timko, M.P.; Zhu, Q.; Fan, L. Analysis of transcriptional and epigenetic changes in hybrid vigor of allopolyploid Brassica napus uncovers key roles for small RNAs. Plant J. 2017, 91, 874–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhang, Z.; Ji, H.; Xu, S.; Yang, Y.; Jia, C.; Zhu, F.; Meng, Q.; Sun, R.; Zhang, Z.; et al. Transcriptome profiles of F1 hybrids (Acanthopagrus schlegelii ♂ × Pagrus major ♀) and parents reveal hybrid effects on individual development. Aquac. Res. 2020, 51, 4011–4021. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, H.; Gao, Q.; Wang, L.; Zhang, F.; Siva, V.S.; Zhou, Z.; Song, L.; Zhang, S. Transcriptome analysis of artificial hybrid pufferfish Jiyan-1 and its parental species: Implications for pufferfish heterosis. PLoS ONE 2013, 8, e58453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Guo, C.; Wang, D.; Li, X.F.; Xiao, L.; Zhang, X.; You, X.; Shi, Q.; Hu, G.; Fang, C.; et al. Transcriptome analysis reveals the molecular mechanisms underlying growth superiority in a novel grouper hybrid (Epinephelus fuscogutatus♀ × E. lanceolatus♂). BMC Genet. 2016, 17, 24. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Li, J.; Zhang, J.; Liang, X.; Zhang, X.; Wang, T.; Yin, S. Integrated analysis of transcriptomic, miRNA and proteomic changes of a novel hybrid yellow catfish uncovers key roles for miRNAs in heterosis. Mol. Cell Proteom. 2019, 18, 1437–1453. [Google Scholar] [CrossRef]
- Zheng, G.; Wu, C.; Liu, J.; Chen, J.; Zou, S. Transcriptome analysis provides new insights into the growth superiority of a novel backcross variety, Megalobrama amblycephala ♀ × (M. amblycephala ♀ × Culter alburnus ♂) ♂. Aquaculture 2019, 512, 734317. [Google Scholar] [CrossRef]
- Yuan, Y.; Yuan, Y.; Dai, Y.; Zhang, Z.; Gong, Y.; Yuan, Y. Technical efficiency of different farm sizes for tilapia farming in China. Aquac. Res. 2019, 51, 307–315. [Google Scholar] [CrossRef]
- Wang, J.; Xia, D. Studies on fish heterosis with DNA fingerprinting. Aquac. Res. 2002, 33, 942–947. [Google Scholar] [CrossRef]
- Zhong, H.; Zhang, X.; Xu, Q.; Yan, J.; Han, Z.; Zheng, H.; Xiao, J.; Tang, Z.; Wang, F.; Luo, Y.; et al. Nonadditive and asymmetric allelic expression of growth hormone in hybrid tilapia. Front. Genet. 2019, 10, 961. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, X.; Xu, Q.; Yan, J.; Yu, F.; Wang, F.; Xiao, J.; Luo, Y.; Zhong, H. Nonadditive and allele-specific expression of insulin-like growth factor 1 in Nile tilapia (Oreochromis niloticus, ♀) × blue tilapia (O. aureus, ♂) hybrids. Comp. Biochem. Physiol. Biochem. Mol. Biol. 2019, 232, 93–100. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, Z.; Chen, W.; Li, X.; Zhou, S.; Liang, C.; Li, X.; Yang, B.; Zou, X.; Liu, F.; et al. Integration of mRNA and miRNA profiling reveals the heterosis of three hybrid combinations of Capsicum annuum varieties. GM Cops Food 2021, 12, 224–241. [Google Scholar] [CrossRef] [PubMed]
- Conte, M.A.; Gammerdinger, W.J.; Bartie, K.L.; Penman, D.J.; Kocher, T.D. A high quality assembly of the Nile tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genom. 2017, 18, 341. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 4, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Bougas, B.; Granier, S.; Audet, C.; Bernatchez, L. The transcriptional landscape of cross-specific hybrids and its possible link with growth in brook charr (Salvelinus fontinalis Mitchill). Genetics 2010, 186, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Yoo, M.; Szadkowski, E.; Wendel, J. Homoeolog expression bias and expression level dominance in allopolyploid cotton. Heredity 2013, 110, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Mao, X.; Huang, J.; Yang, D.; Wu, J.; Dong, S.; Lei, K.; Ge, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar] [CrossRef] [Green Version]
- Rapp, R.A.; Udall, J.A.; Wendel, J.F. Genomic expression dominance in allopolyploids. BMC Biol. 2009, 7, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rehmsmeier, M. Fast and effective prediction of microRNA/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Qiu, C.; Tu, J.; Geng, B.; Yang, J.; Jiang, T.; Cui, Q. HMDD v2.0: A database for experimentally supported human microRNA and disease associations. Nucleic Acids Res. 2014, 42, 1070–1074. [Google Scholar] [CrossRef] [Green Version]
- Cao, D.; Wang, J.; Ji, Z.; Shangguan, Y.; Guo, W.; Feng, X.; Xu, K.; Yang, J. Profiling the mRNA and miRNA in peripheral blood mononuclear cells in subjects with active tuberculosis. Infect. Drug Resist. 2020, 13, 4223–4234. [Google Scholar] [CrossRef]
- Tripp-Valdez, M.A.; Cicala, F.; Galindo-Sánchez, C.E.; Chacón-Ponce, K.D.; López-Landavery, E.; Díaz, F.; Re-Araujo, D.; Lafarga-De La Cruz, F. Growth performance and transcriptomic response of warm-acclimated hybrid abalone Haliotis rufescens (♀) × H. corrugata (♂). Mar. Biotechnol. 2021, 23, 62–76. [Google Scholar] [CrossRef]
- Zhou, Y.; Ren, L.; Xiao, J.; Zhong, H.; Wang, J.; Hu, J.; Yu, F.; Tao, M.; Zhang, C.; Liu, Y. Global transcriptional and miRNA insights into bases of heterosis in hybridization of Cyprinidae. Sci. Rep. 2015, 5, 13847. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, U.A.; Yasid, N.A.; Daud, H.M.; Shukor, M.Y. Glyphosate herbicide induces changes in the growth pattern and somatic indices of crossbred red tilapia (O. niloticus × O. mossambicus). Animals 2021, 11, 1209. [Google Scholar] [CrossRef]
- Hulata, G. Genetic manipulations in aquaculture: A review of stock improvement by classical and modern technologies. Genetica 2001, 111, 155–173. [Google Scholar] [CrossRef]
- El-Zaeem, S. Growth comparison of Nile tilapia (Oreochromis niloticus) and blue tilapia, (Oreochromis aureus) as affected by classical and modern breeding methods. Afr. J. Biotechnol. 2011, 56, 12071–12078. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.F.; Zou, L.L.; Xie, J.G.; Huang, Z.C.; Li, Y.W.; Li, A.X. Immune responses of different species of tilapia infected with Streptococcus agalactiae. J. Fish Dis 2013, 36, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Zou, Z.; Li, D.; Zhu, J.; Yue, Y.; Yang, H. Effect of dietary phenylalanine level on growth performance, body composition, and biochemical parameters in plasma of juvenile hybrid tilapia, Oreochromis niloticus × Oreochromis aureus. J. World Aquacult. Soc. 2020, 51, 437–451. [Google Scholar] [CrossRef]
- Rahimnejad, S.; Lee, K. Dietary valine requirement of juvenile red sea bream Pagrus major. Aquaculture 2013, 416–417, 212–218. [Google Scholar] [CrossRef]
- Ren, M.; Liu, B.; Habte-Tsion, H.; Ge, X.; Xie, J.; Zhou, Q.; Liang, H.; Zhao, Z.; Pan, L. Dietary phenylalanine requirement and tyrosine replacement value for phenylalanine of juvenile blunt snout bream, Megalobrama amblycephala. Aquaculture 2015, 442, 51–57. [Google Scholar] [CrossRef]
- Hsieh, S.; Shiau, S. Effects of diets containing different carbohydrates on starved condition in juvenile tilapia Oreochromis niloticus × O. aureus. Fish Sci. 2000, 66, 32–37. [Google Scholar] [CrossRef]
- Zhu, T.; Mai, K.; Xu, W.; Ai, Q. Effect of dietary cholesterol and phospholipids on feed intake, growth performance and cholesterol metabolism in juvenile turbot (Scophthalmus maximus L.). Aquaculture 2018, 495, 443–451. [Google Scholar] [CrossRef]
- Zhu, T.; Corraze, G.; Plagnes-Juan, E.; Skiba-Cassy, S. Cholesterol metabolism regulation mediated by SREBP-2, LXRα and miR-33a in rainbow trout (Oncorhynchus mykiss) both in vivo and in vitro. PLoS ONE 2020, 15, e223813. [Google Scholar] [CrossRef]
- Bar-Zvi, D.; Lupo, O.; Levy, A.A.; Barkai, N. Hybrid vigor: The best of both parents, or a genomic clash. Curr. Opin. Syst. Biol. 2017, 6, 22–27. [Google Scholar] [CrossRef]
- Tirosh, I.; Reikhav, S.; Levy, A.A.; Barkai, N. A yeast hybrid provides insight into the evolution of gene expression regulation. Science 2009, 324, 659–662. [Google Scholar] [CrossRef]
- Gao, K.; Wang, Z.; Qiu, X.; Song, J.; Wang, H.; Zhao, C.; Wang, X.; Chang, Y. Transcriptome analysis of body wall reveals growth difference between the largest and smallest individuals in the pure and hybrid populations of Apostichopus japonicus. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 31, 100591. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Tan, H.; Luo, L.; Cui, J.; Hu, J.; Wang, S.; Liu, Q.; Hu, F.; Tang, C.; et al. Asymmetric expression patterns reveal a strong maternal effect and dosage compensation in polyploid hybrid fish. BMC Genom. 2018, 19, 517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, L.; Li, W.; Tao, M.; Qin, Q.; Luo, J.; Chai, J.; Tang, C.; Xiao, J.; Tang, X.; Lin, G.; et al. Homoeologue expression insights into the basis of growth heterosis at the intersection of ploidy and hybridity in Cyprinidae. Sci. Rep. 2016, 6, 27040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Huang, Y.; Hu, G.; Zhang, X.; Ruan, Z.; Zhao, X.; Guo, C.; Tang, Z.; Li, X.; You, X.; et al. Comparative transcriptomic study of muscle provides new insights into the growth superiority of a novel grouper hybrid. PLoS ONE 2016, 11, e168802. [Google Scholar] [CrossRef] [PubMed]
- Marcon, C.; Paschold, A.; Malik, W.A.; Lithio, A.; Baldauf, J.A.; Altrogge, L.; Opitz, N.; Lanz, C.; Schoof, H.; Nettleton, D.; et al. Stability of single-parent gene expression complementation in maize hybrids upon water deficit stress. Plant Physiol. 2017, 173, 1247–1257. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.C.; Witucka-Wall, H.; Becher, M.; Blacha, A.; Boudichevskaia, A.; Dörmann, P.; Fiehn, O.; Friedel, S.; von Korff, M.; Lisec, J.; et al. Heterosis manifestation during early Arabidopsis seedling development is characterized by intermediate gene expression and enhanced metabolic activity in the hybrids. Plant J. 2012, 71, 669–683. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, J.; Qu, Y.; Peng, R.; Magwanga, R.O.; Liu, F.; Huang, J. Transcriptomic and proteomic analyses of a new cytoplasmic male sterile line with a wild Gossypium bickii genetic background. BMC Genom. 2020, 21, 859. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yao, H.; Chudalayandi, S.; Vaiman, D.; Veitia, R.A. Heterosis. Plant Cell 2010, 7, 2105–2112. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Luo, S.; Li, J.; Zheng, Z.; Du, X.; Deng, Y. Transcriptome analysis of growth heterosis in pearl oyster Pinctada fucata martensii. FEBS Openbio 2018, 8, 1794–1803. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, X.; Xu, Q.; Yan, J.; Yu, F.; Xiao, J.; Guo, Z.; Luo, Y.; Zhong, H. Nonadditive expression of lipid metabolism pathway-related genes in intestine of hybrids of Nile tilapia females (Oreochromis niloticus) and blue tilapia males (Oreochromis aureus). Mol. Biol. Rep. 2019, 46, 425–432. [Google Scholar] [CrossRef]
- Zhou, X.; Chang, Y.; Zhan, Y.; Wang, X.; Lin, K. Integrative mRNA-miRNA interaction analysis associate with immune response of sea cucumber Apostichopus japonicus based on transcriptome database. Fish Shellfish Immun. 2018, 72, 69–76. [Google Scholar] [CrossRef]
- Mennigen, J.A. Micromanaging metabolism—A role for miRNAs in teleost energy metabolism. Comp. Biochem. Physiol. Biochem. Mol. Biol. 2016, 199, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Johnston, I.A.; Lee, H.; Macqueen, D.J.; Paranthaman, K.; Kawashima, C.; Anwar, A.; Kinghorn, J.R.; Dalmay, T. Embryonic temperature affects muscle fibre recruitment in adult zebrafish: Genome-wide changes in gene and microRNA expression associated with the transition from hyperplastic to hypertrophic growth phenotypes. J. Exp. Biol. 2009, 212, 1781–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Hu, H.; Li, R.; Wang, Z.; Qi, J.; Wang, Z. The role of miR-92 in regulating early development and metamorphosis of Japanese flounder Paralichthys olivaceus. Genes Genet. Syst. 2020, 95, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grover, C.E.; Gallagher, J.P.; Szadkowski, E.P.; Yoo, M.J.; Flagel, L.E.; Wendel, J.F. Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 2012, 196, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Bougas, B.; Audet, C.; Bernatchez, L. The influence of parental effects on transcriptomic landscape during early development in brook charr (Salvelinus fontinalis, Mitchill). Heredity 2013, 110, 484–491. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.J. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010, 15, 57–71. [Google Scholar] [CrossRef] [Green Version]
- Greco, M.; Chiappetta, A.; Bruno, L.; Bitonti, M.B. Heterosis: Emerging ideas about hybrid vigour. J. Exp. Bot. 2012, 63, 695–709. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Cui, J.; Song, J.; Wang, H.; Gao, K.; Qiu, X.; Gou, M.; Li, X.; Hu, Z.; Wang, X.; et al. Comparative transcriptome analysis teveals growth-related genes in juvenile Chinese sea cucumber, Russian sea cucumber, and their hybrids. Mar. Biotechnol. 2018, 20, 193–205. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, X.; He, Z.; Zheng, T.; Shao, J. De novo transcriptome analysis reveals insights into different mechanisms of growth and immunity in a Chinese soft-shelled turtle hybrid and the parental varieties. Gene 2017, 605, 54–62. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, Y.; Lu, G.; Huang, T.; Cai, Y. Analysis of cancer-related lncRNAs using gene ontology and KEGG pathways. Artif. Intell. Med. 2017, 76, 27–36. [Google Scholar] [CrossRef]
- White, S.L.; Sakhrani, D.; Danzmann, R.G.; Devlin, R.H. Influence of developmental stage and genotype on liver mRNA levels among wild, domesticated, and hybrid rainbow trout (Oncorhynchus mykiss). BMC Genom. 2013, 14, 673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di, G.; You, W.; Yu, J.; Wang, D.; Ke, C. Genetic changes in muscle protein following hybridization between Haliotis diversicolor reeve Japan and Taiwan populations revealed using a proteomic approach. Proteomics 2013, 13, 845–859. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; You, W.; Luo, X.; Ke, J.; Huang, M.; Guo, Y.; Ke, C. Integrated analysis of mRNA and miRNA changes in two haliotis diversicolor genotypes and their hybrid. Front. Mar. Sci. 2021, 8, 667636. [Google Scholar] [CrossRef]
- Santos, C.A.; Andrade, S.C.S.; Teixeira, A.K.; Farias, F.; Guerrelhas, A.C.; Rocha, J.L.; Freitas, P.D. Transcriptome differential expression analysis reveals the activated genes in Litopenaeus vannamei shrimp families of superior growth performance. Aquaculture 2021, 531, 735871. [Google Scholar] [CrossRef]
- Kültz, D.; Avila, K. Mitogen-activated protein kinases are in vivo transducers of osmosensory signals in fish gill cells. Comp. Biochem. Physiol. Biochem. Mol. Biol. Part B 2001, 129, 821–829. [Google Scholar] [CrossRef]
- Qian, B.; Qi, X.; Bai, Y.; Wu, Y. The p53 signaling pathway of the large yellow croaker (Larimichthys crocea) responds to acute cold stress: Evidence via spatiotemporal expression analysis of p53, p21, MDM2, IGF-1, Gadd45, Fas, and Akt. PeerJ 2020, 8, e10532. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, W.; Chen, B.; Wang, J.; Zou, Z.; Wang, C.; Li, D.; Zhu, J.; Yu, J.; Yang, H. Integration of mRNA and miRNA Profiling Reveals Heterosis in Oreochromis niloticus × O. aureus Hybrid Tilapia. Animals 2022, 12, 640. https://doi.org/10.3390/ani12050640
Xiao W, Chen B, Wang J, Zou Z, Wang C, Li D, Zhu J, Yu J, Yang H. Integration of mRNA and miRNA Profiling Reveals Heterosis in Oreochromis niloticus × O. aureus Hybrid Tilapia. Animals. 2022; 12(5):640. https://doi.org/10.3390/ani12050640
Chicago/Turabian StyleXiao, Wei, Binglin Chen, Jun Wang, Zhiying Zou, Chenghui Wang, Dayu Li, Jinglin Zhu, Jie Yu, and Hong Yang. 2022. "Integration of mRNA and miRNA Profiling Reveals Heterosis in Oreochromis niloticus × O. aureus Hybrid Tilapia" Animals 12, no. 5: 640. https://doi.org/10.3390/ani12050640





