Retrotransposon Insertion Polymorphisms (RIPs) in Pig Coat Color Candidate Genes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Sequence Acquisition for Coat Color Genes
2.3. Structural Variation Prediction, Retrotransposon Annotation, and Insertion Polymorphic Prediction
2.4. Animals for RIPs Verification and Genotyping
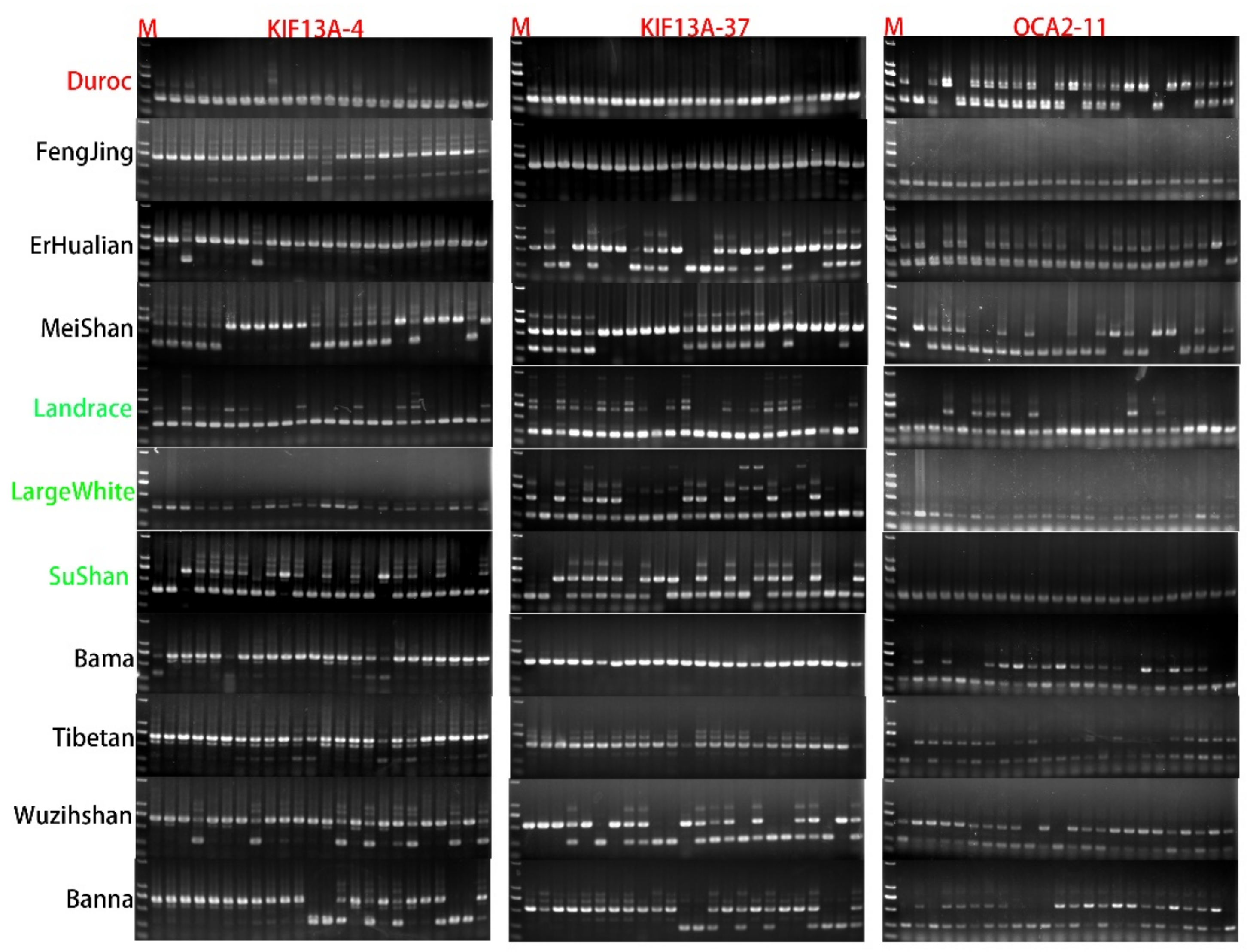
2.5. Samples Collection and PCR Analysis
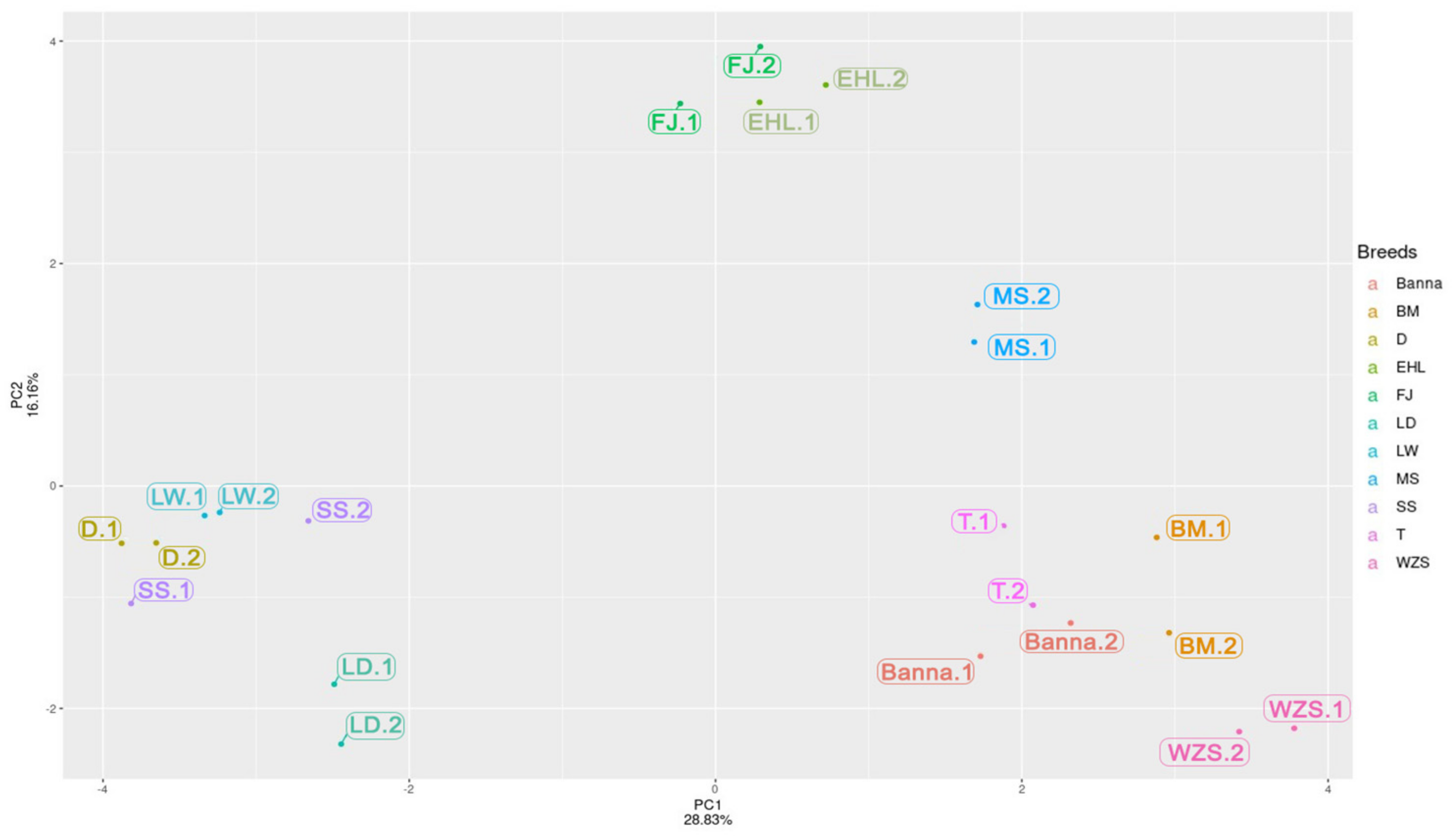
2.6. PCA and Cluster Analysis of the SINE RIPs
3. Results
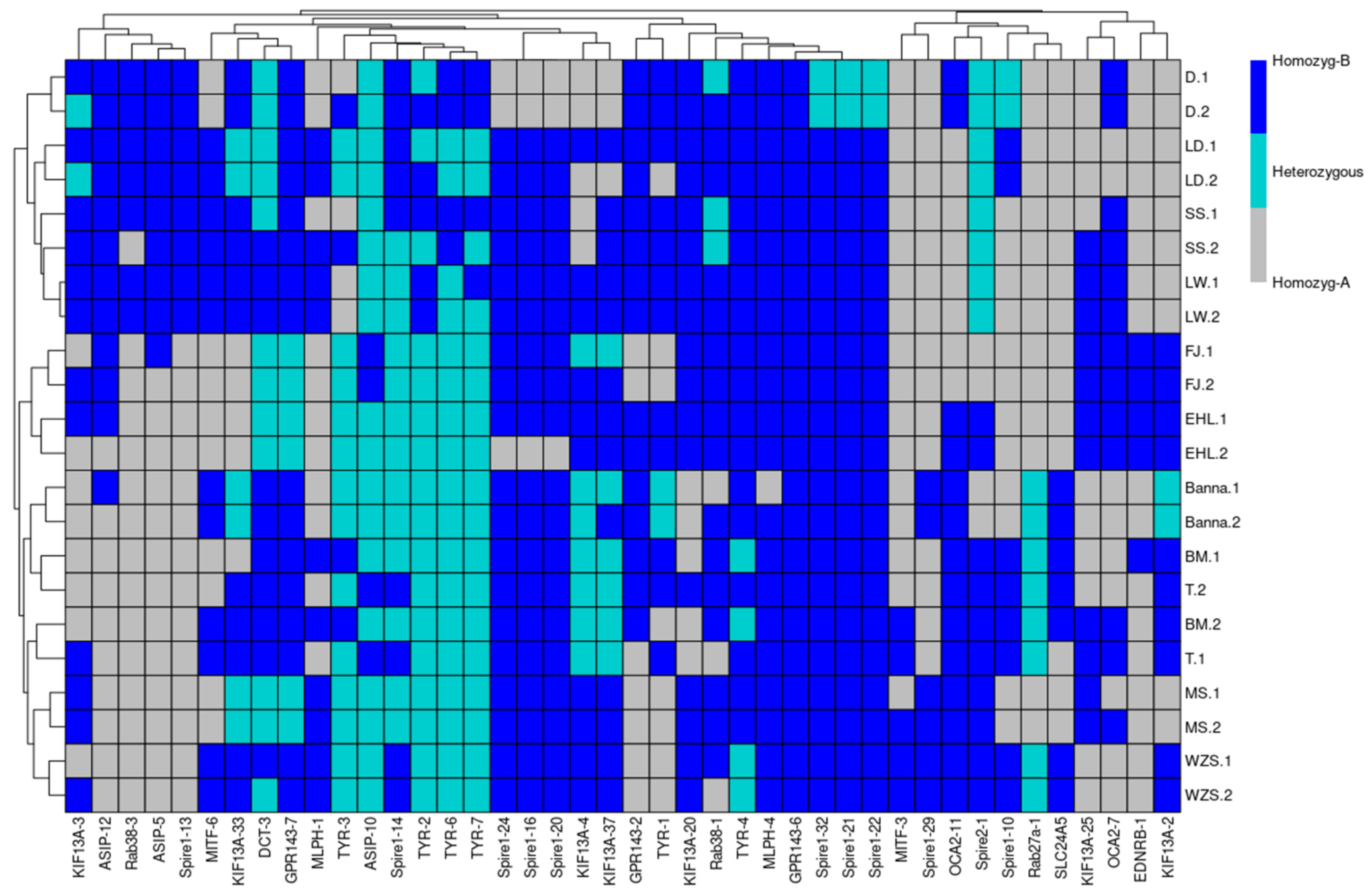
Coat-Color Genes’ SVs Revelation and Detection by Pool-PCR in Different Pig Breeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cuthill, I.C.; Allen, W.L.; Arbuckle, K.; Caspers, B.; Chaplin, G.; Hauber, M.E.; Hill, G.E.; Jablonski, N.G.; Jiggins, C.D.; Kelber, A. The biology of color. Science 2017, 357, 6350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, L.; Sirés-Campos, J.; Raposo, G.; Delevoye, C.; Marks, M.S. Melanosome biogenesis in the pigmentation of mammalian skin. Integr. Comp. Biol. 2021, 61, 1517–1545. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Marks, M.S. Melanosomes—Dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell. Biol. 2007, 8, 786–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreiras, H.; Seabra, M.C.; Barral, D.C. Melanin Transfer in the Epidermis: The Pursuit of Skin Pigmentation Control Mechanisms. Int. J. Mol. Sci. 2021, 22, 4466. [Google Scholar] [CrossRef] [PubMed]
- Benito-Martinez, S.; Salavessa, L.; Raposo, G.; Marks, M.S.; Delevoye, C. Melanin transfer and fate within keratinocytes in human skin pigmentation. Integr. Comp. Biol. 2021, 61, 1546–1555. [Google Scholar] [CrossRef]
- Wiriyasermkul, P.; Moriyama, S.; Nagamori, S. Membrane transport proteins in melanosomes: Regulation of ions for pigmentation. Biochim. Biophys. Acta (BBA)—Biomembr. 2020, 1862, 183318. [Google Scholar] [CrossRef]
- Graham, M.; Tzika, A.C.; Mitchell, S.M.; Liu, X.; Leonhardt, R.M. Repeat domain-associated O-glycans govern PMEL fibrillar sheet architecture. Sci. Rep. 2019, 9, 6101. [Google Scholar] [CrossRef] [Green Version]
- Falletta, P.; Bagnato, P.; Bono, M.; Monticone, M.; Schiaffino, M.V.; Bennett, D.C.; Goding, C.R.; Tacchetti, C.; Valetti, C. Melanosome-autonomous regulation of size and number: The OA 1 receptor sustains PMEL expression. Pigment Cell Melanoma Res. 2014, 27, 565–579. [Google Scholar] [CrossRef]
- Le, L.; Escobar, I.E.; Ho, T.; Lefkovith, A.J.; Latteri, E.; Haltaufderhyde, K.D.; Dennis, M.K.; Plowright, L.; Sviderskaya, E.V.; Bennett, D.C. SLC45A2 protein stability and regulation of melanosome pH determine melanocyte pigmentation. Mol. Biol. Cell. 2020, 31, 2687–2702. [Google Scholar] [CrossRef]
- Yousaf, S.; Sethna, S.; Chaudhary, M.A.; Shaikh, R.S.; Riazuddin, S.; Ahmed, Z.M. Molecular characterization of SLC24A5 variants and evaluation of Nitisinone treatment efficacy in a zebrafish model of OCA6. Pigment Cell Melanoma Res. 2020, 33, 556–565. [Google Scholar] [CrossRef]
- Fukuda, M. Rab GTPases: Key players in melanosome biogenesis, transport, and transfer. Pigment Cell Melanoma Res. 2021, 34, 222–235. [Google Scholar] [CrossRef]
- Alzahofi, N.; Welz, T.; Robinson, C.L.; Page, E.L.; Briggs, D.A.; Stainthorp, A.K.; Reekes, J.; Elbe, D.A.; Straub, F.; Kallemeijn, W.W. Rab27a co-ordinates actin-dependent transport by controlling organelle-associated motors and track assembly proteins. Nat. Commun. 2020, 11, 3495. [Google Scholar] [CrossRef]
- Mort, R.L.; Jackson, I.J.; Patton, E.E. The melanocyte lineage in development and disease. Development 2015, 142, 620–632. [Google Scholar] [CrossRef] [Green Version]
- Pennamen, P.; Le, L.; Tingaud-Sequeira, A.; Fiore, M.; Bauters, A.; Béatrice, N.V.D.; Coste, V.; Bordet, J.; Plaisant, C.; Diallo, M. BLOC1S5 pathogenic variants cause a new type of Hermansky–Pudlak syndrome. Genet. Med. 2020, 22, 1613–1622. [Google Scholar] [CrossRef]
- Xu, X.; Dong, G.; Schmidt-Küntzel, A.; Zhang, X.; Zhuang, Y.; Fang, R.; Sun, X.; Hu, X.; Zhang, T.; Yang, H. The genetics of tiger pelage color variations. Cell Res. 2017, 27, 954–957. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, A.; Pruvost, M.; Reissmann, M.; Benecke, N.; Brockmann, G.A.; Castaños, P.; Cieslak, M.; Lippold, S.; Llorente, L.; Malaspinas, A. Coat color variation at the beginning of horse domestication. Science 2009, 324, 485. [Google Scholar] [CrossRef] [Green Version]
- Corbin, L.J.; Pope, J.; Sanson, J.; Antczak, D.F.; Miller, D.; Sadeghi, R.; Brooks, S.A. An Independent Locus Upstream of ASIP Controls Variation in the Shade of the Bay Coat Colour in Horses. Genes 2020, 11, 606. [Google Scholar] [CrossRef]
- Hédan, B.; Cadieu, E.; Botherel, N.; Dufaure de Citres, C.; Letko, A.; Rimbault, M.; Drögemüller, C.; Jagannathan, V.; Derrien, T.; Schmutz, S. Identification of a missense variant in MFSD12 involved in dilution of Phaeomelanin leading to white or cream coat color in dogs. Genes 2019, 10, 386. [Google Scholar] [CrossRef] [Green Version]
- Schmutz, S.M. Genetics of coat color in cattle. In Bovine Genomics; Wiley Publication: Hoboken, NJ, USA, 2012; pp. 20–33. [Google Scholar]
- Trigo, B.B.; Utsunomiya, A.T.H.; Fortunato, A.A.; Milanesi, M.; Torrecilha, R.B.P.; Lamb, H.; Nguyen, L.; Ross, E.M.; Hayes, B.; Padula, R.C.M. Variants at the ASIP locus contribute to coat color darkening in Nellore cattle. Genet. Sel. Evol. 2021, 53, 40. [Google Scholar] [CrossRef]
- Demars, J.; Iannuccelli, N.; Utzeri, V.J.; Auvinet, G.; Riquet, J.; Fontanesi, L.; Allain, D. New insights into the melanophilin (MLPH) gene affecting coat color dilution in rabbits. Genes 2018, 9, 430. [Google Scholar] [CrossRef] [Green Version]
- Manakhov, A.D.; Andreeva, T.V.; Trapezov, O.V.; Kolchanov, N.A.; Rogaev, E.I. Genome analysis identifies the mutant genes for common industrial Silverblue and Hedlund white coat colours in American mink. Sci. Rep. 2019, 9, 4581. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.L.; Kozysa, A.; Kharlamova, A.V.; Gulevich, R.G.; Perelman, P.L.; Fong, H.; Vladimirova, A.V.; Oskina, I.N.; Trut, L.N.; Kukekova, A.V. Platinum coat color in red fox (Vulpes vulpes) is caused by a mutation in an autosomal copy of KIT. Anim. Genet. 2015, 46, 190–199. [Google Scholar] [CrossRef]
- Du, Z.; Huang, K.; Zhao, J.; Song, X.; Xing, X.; Wu, Q.; Zhang, L.; Xu, C. Comparative transcriptome analysis of raccoon dog skin to determine melanin content in hair and melanin distribution in skin. Sci. Rep. 2017, 7, 40903. [Google Scholar] [CrossRef]
- Kijas, J.; Wales, R.; Törnsten, A.; Chardon, P.; Moller, M.; Andersson, L. Melanocortin receptor 1 (MC1R) mutations and coat color in pigs. Genetics 1998, 150, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Deng, Z.; Huang, M.; Hou, Y.; Zhang, H.; Chen, H.; Ren, J. Whole-genome resequencing identifies KIT new alleles that affect coat color phenotypes in pigs. Front. Genet. 2019, 10, 218. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, F.; Kong, X.; Tan, B.; Li, Y.; Duan, Y.; François, B.; Chien-An, A.; Yin, Y. Signaling pathways related to protein synthesis and amino acid concentration in pig skeletal muscles depend on the dietary protein level, genotype and developmental stages. PLoS ONE 2015, 10, e0138277. [Google Scholar] [CrossRef] [PubMed]
- Whitelaw, E.; Martin, D.I.K. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat. Genet. 2001, 27, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [Green Version]
- Blumenstiel, J.P. Birth, school, work, death, and resurrection: The life stages and dynamics of transposable element proliferation. Genes 2019, 10, 336. [Google Scholar] [CrossRef] [Green Version]
- Platt, R.N.; Vandewege, M.W.; Ray, D.A. Mammalian transposable elements and their impacts on genome evolution. Chromosom. Res. 2018, 26, 25–43. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Wang, W.; Wang, X.; Shen, D.; Wang, S.; Wang, Y.; Gao, B.; Wimmers, K.; Mao, J.; Li, K. Retrotransposons evolution and impact on lncRNA and protein coding genes in pigs. Mob. DNA 2019, 10, 19. [Google Scholar] [CrossRef]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Lancíková, V.; Žiarovská, J. Inter-retrotransposon amplified polymorphism markers revealed long terminal repeat retrotransposon insertion polymorphism in flax cultivated on the experimental fields around Chernobyl. J. Environ. Sci. Health Part. A 2020, 55, 957–963. [Google Scholar] [CrossRef]
- Hirata, C.; Waki, T.; Shimomura, K.; Wada, T.; Tanaka, S.; Ikegami, H.; Uchimura, Y.; Hirashima, K.; Nakazawa, Y.; Okada, K.; et al. DNA markers based on retrotransposon insertion polymorphisms can detect short DNA fragments for strawberry cultivar identification. Breed. Sci. 2020, 70, 19116. [Google Scholar] [CrossRef] [Green Version]
- Meng, Y.; Su, W.; Ma, Y.; Liu, L.; Gu, X.; Wu, D.; Shu, X.; Lai, Q.; Tang, Y.; Wu, L.; et al. Assessment of genetic diversity and variety identification based on developed retrotransposon-based insertion polymorphism (RBIP) markers in sweet potato (Ipomoea batatas (L.) Lam.). Sci. Rep. 2021, 11, 17116. [Google Scholar] [CrossRef]
- Pelsy, F.; Bevilacqua, L.; Blanc, S.; Merdinoglu, D. A molecular marker set combining a retrotransposon insertion and SSR polymorphisms is useful for assessing diversity in Vitis. OENO One 2021, 55, 403–414. [Google Scholar] [CrossRef]
- Borges-Monroy, R.; Chu, C.; Dias, C.; Choi, J.; Lee, S.; Gao, Y.; Shin, T.; Park, P.J.; Walsh, C.A.; Eunjung Alice Lee, E.A. Whole-genome analysis of de novo and polymorphic retrotransposon insertions in Autism Spectrum Disorder. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kalla, S.E.; Moghadam, H.K.; Tomlinson, M.; Seebald, A.; Allen, J.J.; Whitney, J.; Choi, J.D.; Sutter, N.B. Polymorphic SINEC_Cf Retrotransposons in the Genome of the Dog (Canis familiaris). bioRxiv 2020. [Google Scholar] [CrossRef]
- Choi, J.D.; Del Pinto, L.A.; Sutter, N.B. SINE Retrotransposons Import Polyadenylation Signals to 3′UTRs in Dog (Canis familiaris). bioRxiv 2020. [Google Scholar] [CrossRef]
- Ruggiero, R.P.; Bourgeois, Y.; Boissinot, S. LINE insertion polymorphisms are abundant but at low frequencies across populations of Anolis carolinensis. Front. Genet. 2017, 8, 44. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, C.; Wang, X.; Zhang, L.; Shen, D.; Wang, S.; Gao, B.; Mao, J.; Song, C. Development of Molecular Markers Based on the L1 Retrotransposon Insertion Polymorphisms in Pigs (Sus scrofa) and Their Association with Economic Traits. Russ. J. Genet. 2020, 56, 183–191. [Google Scholar] [CrossRef]
- Zheng, Y.; Cai, C.; Wei, C.; Wang, X.; Wei, W.; Bo, G.; Wimmers, K.; Mao, J.; Song, C. Two new SINE insertion polymorphisms in pig Vertnin (VRTN) gene revealed by comparative genomic alignment. J. Integ. Agri. 2020, 19, 2514–2522. [Google Scholar] [CrossRef]
- Chen, C.; Zheng, Y.; Wang, M.; Murani, E.; D’Alessandro, E.; Moawad, A.S.; Wang, X.; Wimmers, K.; Song, C. SINE Insertion in the Intron of Pig GHR May Decrease Its Expression by Acting as a Repressor. Animals 2021, 11, 1871. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, D. Repeat Masker. Biotech. Softw. Internet Rep. 2000, 1, 36–39. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps, R Packag v. 16; R Foundation for Statistical Computing: Nashville, TN, USA, 2012. [Google Scholar]
- Zhang, Z.D.; Paccanaro, A.; Fu, Y.; Weussnab, S.; Weng, Z.; Chang, J.; Snyder, M.; Merk, B. Statistical analysis of the genomic distribution and correlation of regulatory elements in the ENCODE regions. Genome Res. 2007, 17, 787–797. [Google Scholar] [CrossRef] [Green Version]
- Zhou, L.; Zhao, W.; Fu, Y.; Fang, X.; Ren, S.; Ren, J. Genome-wide detection of genetic loci and candidate genes for teat number and body conformation traits at birth in Chinese Sushan pigs. Anim. Genet. 2019, 50, 753–756. [Google Scholar] [CrossRef]
- Jia, Q.; Cao, C.; Tang, H.; Ying, Z.; Qian, Z.; Xiao, W.; Rui, Z.; Xiao, W.; Ai, L.; Hong, W.; et al. A 2-bp insertion (c. 67_68insCC) in MC1R causes recessive white coat color in Bama miniature pigs. J. Genet. Genom. 2017, 44, 215–217. [Google Scholar] [CrossRef]
- Weissensteiner, M.H.; Bunikis, I.; Catalán, A.; Francoijs, K.J.; Knief, U.; Heim, W.; Peona, V.; Pophaly, S.D.; Sedlazeck, F.J.; Suh, A.; et al. Discovery and population genomics of structural variation in a songbird genus. Nat. Commun. 2020, 11, 3403. [Google Scholar] [CrossRef]
- Chiang, C.; Scott, A.J.; Davis, J.R.; Tsang, E.K.; Li, X.; Kim, Y.; Hadzic, T.; Damani, F.N.; Ganel, L.; GTEx Consortium; et al. The impact of structural variation on human gene expression. Nat. Genet. 2017, 49, 692–699. [Google Scholar] [CrossRef] [Green Version]
- Yang, N.; Zhao, B.; Chen, Y.; D’Alessandro, E.; Chen, C.; Ji, T.; Wu, X.; Song, C. Distinct retrotransposon evolution profile in the genome of rabbit (Oryctolagus cuniculus). Genome Biol. Evol. 2021, 13, evab168. [Google Scholar]
- Liang, D.; Zhao, P.; Si, J.; Fang, L.; Pairo-Castineira, E.; Hu, X.; Xu, Q.; Hou, Y.; Gong, Y.; Liang, Z.; et al. Genomic analysis revealed a convergent evolution of LINE-1 in coat color: A case study in water buffaloes (Bubalus bubalis). Mol. Biol. Evol. 2021, 38, 1122–1136. [Google Scholar] [CrossRef]
- Li, J.; Davis, B.W.; Jern, P.; Dorshorst, B.J.; Siegel, P.B.; Andersson, L. Characterization of the endogenous retrovirus insertion in CYP19A1 associated with henny feathering in chicken. Mob. DNA 2019, 10, 38. [Google Scholar] [CrossRef]
- David, V.A.; Menotti-Raymond, M.; Wallace, A.C.; Roelke, M.; Kehler, J.; Leighty, R.; Eizirik, E.; Hannah, S.S.; Nelson, G.; Schäffer, A.A.; et al. Endogenous retrovirus insertion in the KIT oncogene determines white and white spotting in domestic cats. G3 Genes. Genomes Genet. 2014, 4, 1881–1891. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Y.; Chen, Z.X.; Murani, E.; D’Alessamdro, E.; An, Y.L.; Chen, C.; Li, K.; Galeano, G.; Wimmers, K.; Song, C.Y. A 192 bp ERV fragment insertion in the first intron of porcine TLR6 may act as an enhancer associated with the increased expressions of TLR6 and TLR1. Mob. DNA 2021, 12, 20. [Google Scholar] [CrossRef]
- Chen, C.; D’Alessandro, E.; Murani, E.; Zheng, Y.; Domenico, G.; Yang, N.S.; Wang, X.Y.; Gao, B.; Li, K.; Wimmer, K.; et al. SINE jumping contributes to large-scale polymorphisms in the pig genomes. Mob. DNA 2021, 12, 17. [Google Scholar] [CrossRef]
- Margeta, V.; Budimir, K.; Kralik, G.; Radišić, Ž. Genetic basis of pig coat color. Krmiva Časopis Hranidbi Zivotinj. Proizv. Tehnol. Krme 2011, 53, 85–92. [Google Scholar]

| Gene Name | Predicted RIPs | Confirmed RIPs |
|---|---|---|
| ASIP | 14 | 2 |
| KIT | 8 | 0 |
| MITF | 12 | 2 |
| PMEL | 1 | 0 |
| GPR143 | 7 | 3 |
| TYR | 7 | 6 |
| EDNRB | 2 | 1 |
| TYRP1 | 0 | 0 |
| MLPH | 11 | 3 |
| DCT | 3 | 1 |
| OCA2 | 10 | 2 |
| SLC24A5 | 1 | 1 |
| SLC45A2 | 9 | 0 |
| Rab27a | 6 | 1 |
| Rab32 | 2 | 0 |
| Rab38 | 4 | 2 |
| KIF13A | 37 | 7 |
| Spire2 | 3 | 1 |
| Spire1 | 30 | 10 |
| MC1R | 0 | 0 |
| Total Numbers | 167 | 42 |
| Rip-Sites | Insertion Breeds | Deletion Breeds | Chr | Begin | End | Gene Struture | TE-Type | Length (bp) |
|---|---|---|---|---|---|---|---|---|
| ASIP-12 | Duroc, Berkshire | The rest of the species | 17 | 37,614,914 | 37,615,246 | Intron-1 | SINEA | 332 |
| GPR143-7 | The rest of the species | Landrace, LargeWhite, Crossbred, Wuzhishan, Bama | X | 6,248,783 | 6,249,085 | Intron-9 | SINEA | 300 |
| MITF-6 | Largewhite, Berkshire | The rest of the species | 13 | 51,175,504 | 51,175,505 | Intron-1 | SINEB | 94 |
| KIF13A-4 | The rest of the species | Berkshire, Crossbred, LargeWhite, D-Ninghe, Hampshire, Landrace, Pietrain | 7 | 13,691,836 | 13,692,190 | Intron-1 | SINEA | 354 |
| KIF13A-37 | Bamei, Wuzhishan, Bama, Tibetan, Rongchang | The rest of the species | 7 | 13,497,155 | 13,497,156 | 3′flank | ERV III | 282 |
| OCA2-11 | Wuzhishan, Jinhua, Rongchang, Hampshire | The rest of the species | 15 | 56,806,804 | 56,806,805 | Intron-2 | SINEA | 339 |
| Rab27a-1 | Bama, Tibetan, Bamei | The rest of the species | 1 | 116,538,879 | 116,538,880 | Intron-1 | SINEA | 326 |
| Rab38-3 | LargeWhite, Landrace, Hampshire, Berkshire | The rest of the species | 9 | 21,583,690 | 21,583,691 | Intron-2 | SINEA | 293 |
| SLC24A5-1 | Bama | The rest of the species | 1 | 123,633,504 | 123,633,505 | 3′flank | SINEA | 310 |
| Spire1-13 | The rest of the species | Bamei, MS, Landrace | 6 | 96,966,838 | 96,967,753 | Intron-2 | SINEA | 370 |
| Spire2-1 | Duroc, Tibetan, Largewhite, D-Ninghe, Crossbred, Pietrain | The rest of the species | 6 | 248,403 | 248,980 | 5′flank | SINEA | 577 |
| Asia-12 | GPR143-7 | MITF-6 | KIF13A-4 | KIF13A-37 | OCA2-11 | Rab27a-1 | Rab38 | SLC24A5-1 | Spire1-13 | Spire2-1 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Duro | +/+ | 6 | 5 | 0 | 0 | 0 | 6 | 0 | 5 | 0 | 0 | 0 |
| +/− | 12 | 17 | 0 | 5 | 0 | 14 | 0 | 13 | 0 | 23 | 24 | |
| −/− | 6 | 2 | 24 | 19 | 24 | 4 | 24 | 6 | 24 | 1 | 0 | |
| Fengjing | +/+ | 1 | 24 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 0 |
| +/− | 14 | 0 | 24 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| −/− | 9 | 0 | 0 | 0 | 0 | 24 | 24 | 24 | 24 | 24 | 24 | |
| ErHualian | +/+ | 0 | 24 | 0 | 22 | 9 | 1 | 0 | 0 | 0 | 0 | 5 |
| +/− | 0 | 0 | 0 | 2 | 11 | 20 | 0 | 0 | 0 | 0 | 17 | |
| −/− | 24 | 0 | 24 | 0 | 4 | 3 | 24 | 24 | 24 | 24 | 2 | |
| Meishan | +/+ | 0 | 24 | 0 | 11 | 11 | 4 | 0 | 0 | 0 | 0 | 0 |
| +/− | 0 | 0 | 0 | 13 | 13 | 10 | 0 | 0 | 0 | 0 | 0 | |
| −/− | 24 | 0 | 24 | 0 | 0 | 10 | 24 | 24 | 24 | 24 | 24 | |
| Landrace | +/+ | 7 | 9 | 7 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 |
| +/− | 15 | 15 | 10 | 12 | 16 | 7 | 0 | 17 | 0 | 20 | 24 | |
| −/− | 2 | 0 | 7 | 12 | 8 | 17 | 24 | 4 | 24 | 4 | 0 | |
| LargeWhite | +/+ | 3 | 6 | 16 | 0 | 0 | 0 | 0 | 8 | 0 | 0 | 0 |
| +/− | 6 | 0 | 8 | 0 | 16 | 24 | 0 | 16 | 0 | 22 | 24 | |
| −/− | 15 | 18 | 0 | 24 | 8 | 0 | 24 | 0 | 24 | 2 | 0 | |
| SuShan | +/+ | 4 | 5 | 16 | 3 | 3 | 0 | 0 | 11 | 0 | 0 | 0 |
| +/− | 11 | 13 | 8 | 12 | 12 | 0 | 0 | 11 | 0 | 14 | 24 | |
| −/− | 9 | 6 | 0 | 9 | 9 | 24 | 24 | 2 | 24 | 7 | 0 | |
| Bama | +/+ | 0 | 0 | 3 | 19 | 24 | 0 | 13 | 0 | 4 | 0 | 0 |
| +/− | 0 | 8 | 4 | 5 | 0 | 17 | 11 | 0 | 7 | 0 | 7 | |
| −/− | 24 | 16 | 17 | 0 | 0 | 7 | 0 | 24 | 13 | 24 | 17 | |
| Tibetan | +/+ | 0 | 4 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 1 |
| +/− | 0 | 18 | 22 | 24 | 18 | 23 | 4 | 0 | 7 | 0 | 15 | |
| −/− | 24 | 2 | 2 | 0 | 6 | 1 | 7 | 24 | 14 | 24 | 8 | |
| Wuzhishan | +/+ | 0 | 4 | 2 | 15 | 7 | 0 | 18 | 0 | 0 | 0 | 1 |
| +/− | 0 | 14 | 3 | 9 | 10 | 22 | 3 | 0 | 16 | 0 | 12 | |
| −/− | 24 | 6 | 19 | 0 | 7 | 2 | 3 | 24 | 8 | 24 | 11 | |
| Banna | +/+ | 0 | 6 | 1 | 16 | 15 | 0 | 18 | 0 | 1 | 0 | 0 |
| +/− | 4 | 12 | 7 | 4 | 5 | 18 | 5 | 0 | 8 | 0 | 10 | |
| −/− | 20 | 6 | 16 | 4 | 4 | 6 | 1 | 24 | 15 | 24 | 14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Z.; D’Alessandro, E.; Zheng, Y.; Wang, M.; Chen, C.; Wang, X.; Song, C. Retrotransposon Insertion Polymorphisms (RIPs) in Pig Coat Color Candidate Genes. Animals 2022, 12, 969. https://doi.org/10.3390/ani12080969
Du Z, D’Alessandro E, Zheng Y, Wang M, Chen C, Wang X, Song C. Retrotransposon Insertion Polymorphisms (RIPs) in Pig Coat Color Candidate Genes. Animals. 2022; 12(8):969. https://doi.org/10.3390/ani12080969
Chicago/Turabian StyleDu, Zhanyu, Enrico D’Alessandro, Yao Zheng, Mengli Wang, Cai Chen, Xiaoyan Wang, and Chengyi Song. 2022. "Retrotransposon Insertion Polymorphisms (RIPs) in Pig Coat Color Candidate Genes" Animals 12, no. 8: 969. https://doi.org/10.3390/ani12080969
APA StyleDu, Z., D’Alessandro, E., Zheng, Y., Wang, M., Chen, C., Wang, X., & Song, C. (2022). Retrotransposon Insertion Polymorphisms (RIPs) in Pig Coat Color Candidate Genes. Animals, 12(8), 969. https://doi.org/10.3390/ani12080969








