Occurrence and Molecular Characteristics of Microsporidia in Captive Red Pandas (Ailurus fulgens) in China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
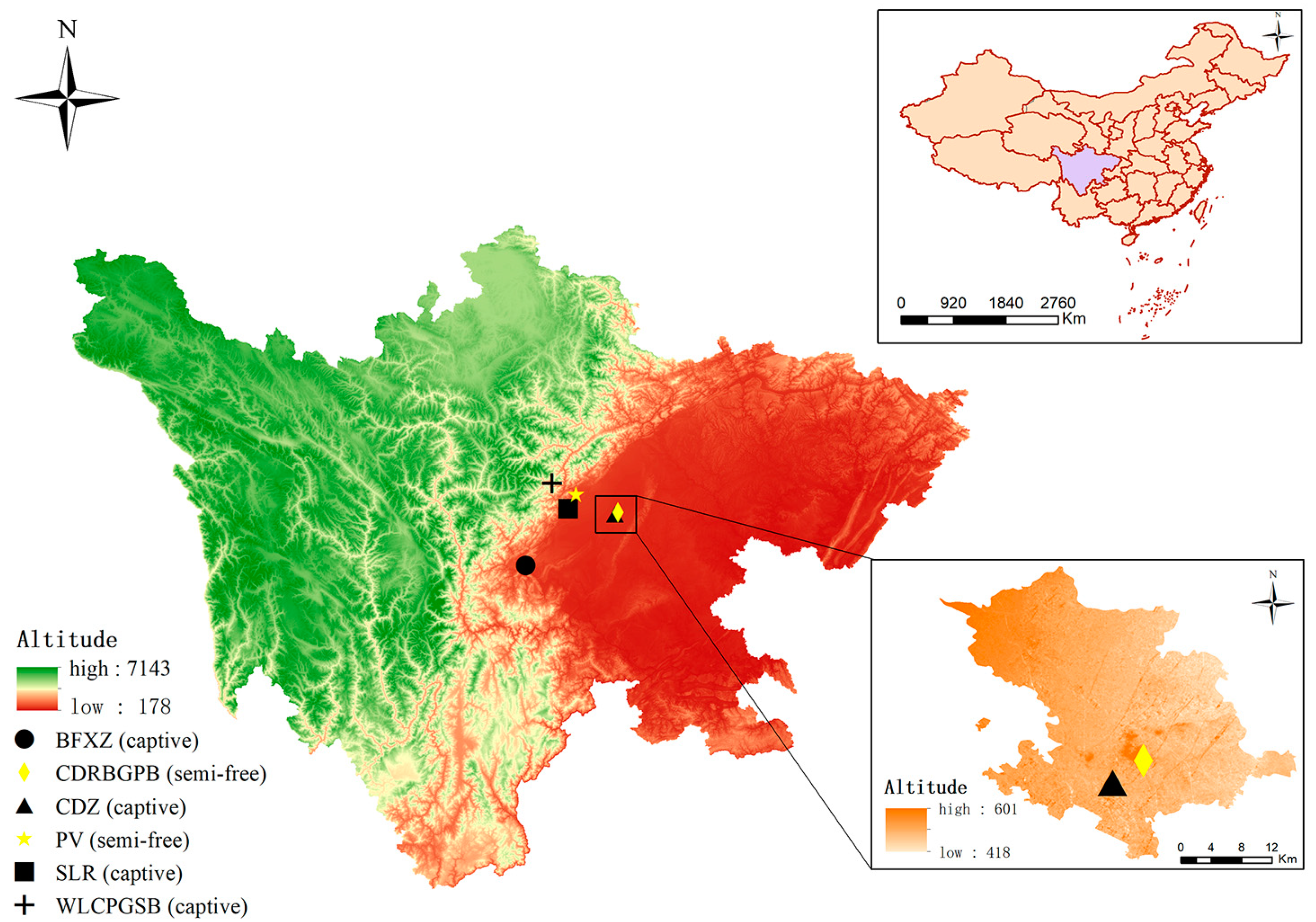
2.2. Sample Collection
2.3. DNA Isolation and PCR Amplification
2.4. Sequencing and Phylogenetic Analysis
2.5. Statistics Analyses
2.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Prevalence of Microsporidia in Fecal Samples
3.2. Genotypes of E. bieneusi and Homology Comparison
3.3. Phylogenetic Relationship of E. bieneusi and Encephalitozoon spp.
3.4. Multi-Locus Sequence Typing of E. bieneusi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, B.; Pan, G.; Weiss, L.M. Microsporidiosis in Humans. Clin. Microbiol. Rev. 2021, 34, e0001020. [Google Scholar] [CrossRef] [PubMed]
- Santín, M.; Fayer, R. Microsporidiosis: Enterocytozoon bieneusi in domesticated and wild animals. Res. Vet. Sci. 2011, 90, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Theng, J.T.; Li, L.; Tan, D.T. Microsporidial keratoconjunctivitis in healthy individuals: A case series. Ophthalmology 2003, 110, 1420–1425. [Google Scholar] [CrossRef]
- Mathis, A.; Weber, R.; Deplazes, P. Zoonotic potential of the microsporidia. Clin. Microbiol. Rev. 2005, 18, 423–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.S.; Wang, R.J.; Fan, X.C.; Liu, T.L.; Zhang, L.X.; Zhao, G.H. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop. 2018, 183, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Koehler, A.V.; Wang, T.; Gasser, R.B. Enterocytozoon bieneusi of animals-with an ’Australian twist’. Adv. Parasitol. 2021, 111, 1–73. [Google Scholar] [CrossRef]
- Li, W.; Feng, Y.; Zhang, L.; Xiao, L. Potential impacts of host specificity on zoonotic or interspecies transmission of Enterocytozoon bieneusi. Infect. Genet. Evol. 2019, 75, 104033. [Google Scholar] [CrossRef]
- Liu, H.; Ni, H.; Xu, J.; Wang, R.; Li, Y.; Shen, Y.; Cao, J.; Yin, J. Genotyping and zoonotic potential of Cryptosporidium and Enterocytozoon bieneusi in pigs transported across regions in China. Microb. Pathog. 2021, 154, 104823. [Google Scholar] [CrossRef]
- Yu, Z.; Wen, X.; Huang, X.; Yang, R.; Guo, Y.; Feng, Y.; Xiao, L.; Li, N. Molecular characterization and zoonotic potential of Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium sp. in farmed masked palm civets (Paguma larvata) in southern China. Parasites Vectors 2020, 13, 403. [Google Scholar] [CrossRef]
- Didier, E.S.; Vossbrinck, C.R.; Baker, M.D.; Rogers, L.B.; Bertucci, D.C.; Shadduck, J.A. Identification and characterization of three Encephalitozoon cuniculi strains. Parasitology 1995, 111 Pt 4, 411–421. [Google Scholar] [CrossRef]
- Talabani, H.; Sarfati, C.; Pillebout, E.; van Gool, T.; Derouin, F.; Menotti, J. Disseminated infection with a new genovar of Encephalitozoon cuniculi in a renal transplant recipient. J. Clin. Microbiol. 2010, 48, 2651–2653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathis, A.; Tanner, I.; Weber, R.; Deplazes, P. Genetic and phenotypic intraspecific variation in the microsporidian Encephalitozoon hellem. Int. J. Parasitol. 1999, 29, 767–770. [Google Scholar] [CrossRef] [PubMed]
- Liguory, O.; Fournier, S.; Sarfati, C.; Derouin, F.; Molina, J.M. Genetic homology among thirteen Encephalitozoon intestinalis isolates obtained from human immunodeficiency virus-infected patients with intestinal microsporidiosis. J. Clin. Microbiol. 2000, 38, 2389–2391. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Feng, Z.; Wang, Z.; Hu, J. Current distribution, status and conservation of wild red pandas Ailurus fulgens in China. Biol. Conserv. 1999, 89, 285–291. [Google Scholar] [CrossRef]
- Deng, L.; Chai, Y.; Xiang, L.; Wang, W.; Zhou, Z.; Liu, H.; Zhong, Z.; Fu, H.; Peng, G. First identification and genotyping of Enterocytozoon bieneusi and Encephalitozoon spp. in pet rabbits in China. BMC Vet. Res. 2020, 16, 212. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Xia, W.; Li, W.; Ping, J.; Ding, S.; Liu, H. The prevalence of microsporidia in China: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 3174. [Google Scholar] [CrossRef]
- Zhang, Y.; Mi, R.; Yang, L.; Gong, H.; Xu, C.; Feng, Y.; Chen, X.; Huang, Y.; Han, X.; Chen, Z. Wildlife Is a Potential Source of Human Infections of Enterocytozoon bieneusi and Giardia duodenalis in Southeastern China. Front. Microbiol. 2021, 12, 692837. [Google Scholar] [CrossRef]
- Ghebremichael, S.T.; Meng, X.; Wei, J.; Yang, Y.; Huang, Q.; Luo, L.; Xiang, H.; Chen, J.; Abo-Kadoum, M.A.; Li, T.; et al. Prevalence and genotyping distribution of Enterocytozoon bieneusi in diarrheic pigs in Chongqing and Sichuan provinces, China. Front. Microbiol. 2022, 13, 1025613. [Google Scholar] [CrossRef]
- Tian, G.R.; Zhao, G.H.; Du, S.Z.; Hu, X.F.; Wang, H.B.; Zhang, L.X.; Yu, S.K. First report of Enterocytozoon bieneusi from giant pandas (Ailuropoda melanoleuca) and red pandas (Ailurus fulgens) in China. Infect. Genet. Evol. 2015, 34, 32–35. [Google Scholar] [CrossRef]
- Li, W.; Deng, L.; Yu, X.; Zhong, Z.; Wang, Q.; Liu, X.; Niu, L.; Xie, N.; Deng, J.; Lei, S.; et al. Multilocus genotypes and broad host-range of Enterocytozoon bieneusi in captive wildlife at zoological gardens in China. Parasites Vectors 2016, 9, 395. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.S.; Daszak, P.; Huang, H.L.; Yang, G.Y.; Kilpatrick, A.M.; Zhang, S. Parasite threat to panda conservation. EcoHealth 2008, 5, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, I.M.; Fayer, R.; Lal, A.A.; Trout, J.M.; Schaefer, F.W., 3rd; Xiao, L. Molecular characterization of microsporidia indicates that wild mammals Harbor host-adapted Enterocytozoon spp. as well as human-pathogenic Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2003, 69, 4495–4501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Y.; Li, N.; Dearen, T.; Lobo, M.L.; Matos, O.; Cama, V.; Xiao, L. Development of a multilocus sequence typing tool for high-resolution genotyping of Enterocytozoon bieneusi. Appl. Environ. Microbiol. 2011, 77, 4822–4828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katzwinkel-Wladarsch, S.; Lieb, M.; Helse, W.; Löscher, T.; Rinder, H. Direct amplification and species determination of microsporidian DNA from stool specimens. Trop. Med. Int. Health TM IH 1996, 1, 373–378. [Google Scholar] [CrossRef]
- Mynářová, A.; Foitová, I.; Kváč, M.; Květoňová, D.; Rost, M.; Morrogh-Bernard, H.; Nurcahyo, W.; Nguyen, C.; Supriyadi, S.; Sak, B. Prevalence of Cryptosporidium spp., Enterocytozoon bieneusi, Encephalitozoon spp. and Giardia intestinalis in Wild, Semi-Wild and Captive Orangutans (Pongo abelii and Pongo pygmaeus) on Sumatra and Borneo, Indonesia. PLoS ONE 2016, 11, e0152771. [Google Scholar] [CrossRef] [Green Version]
- Karim, M.R.; Wang, R.; Dong, H.; Zhang, L.; Li, J.; Zhang, S.; Rume, F.I.; Qi, M.; Jian, F.; Sun, M.; et al. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl. Environ. Microbiol. 2014, 80, 1893–1898. [Google Scholar] [CrossRef] [Green Version]
- Leśniańska, K.; Perec-Matysiak, A. Wildlife as an environmental reservoir of Enterocytozoon bieneusi (Microsporidia)—Analyses of data based on molecular methods. Ann. Parasitol. 2017, 63, 265–281. [Google Scholar]
- Xu, C.; Ma, X.; Zhang, H.; Zhang, X.X.; Zhao, J.P.; Ba, H.X.; Rui, D.; Xing, X.M.; Wang, Q.K.; Zhao, Q. Prevalence, risk factors and molecular characterization of Enterocytozoon bieneusi in raccoon dogs (Nyctereutes procyonoides) in five provinces of Northern China. Acta Trop. 2016, 161, 68–72. [Google Scholar] [CrossRef]
- Yang, Y.; Lin, Y.; Li, Q.; Zhang, S.; Tao, W.; Wan, Q.; Jiang, Y.; Li, W. Widespread presence of human-pathogenic Enterocytozoon bieneusi genotype D in farmed foxes (Vulpes vulpes) and raccoon dogs (Nyctereutes procyonoides) in China: First identification and zoonotic concern. Parasitol. Res. 2015, 114, 4341–4348. [Google Scholar] [CrossRef]
- Zhang, X.X.; Cong, W.; Liu, G.H.; Ni, X.T.; Ma, J.G.; Zheng, W.B.; Zhao, Q.; Zhu, X.Q. Prevalence and genotypes of Enterocytozoon bieneusi in sika deer in Jilin province, Northeastern China. Acta Parasitol. 2016, 61, 382–388. [Google Scholar] [CrossRef]
- Su, B.; Fu, Y.; Wang, Y.; Jin, L.; Chakraborty, R. Genetic diversity and population history of the red panda (Ailurus fulgens) as inferred from mitochondrial DNA sequence variations. Mol. Biol. Evol. 2001, 18, 1070–1076. [Google Scholar] [CrossRef] [Green Version]
- Javanmard, E.; Nemati, S.; Sharifdini, M.; Rostami, A.; Mirjalali, H.; Zali, M.R. The First Report and Molecular Analysis of Enterocytozoon bieneusi from Raccoon (Procyon lotor) in North of Iran. J. Eukaryot. Microbiol. 2020, 67, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Wu, Y.; Li, T.; Cao, J.; Wang, J.; Hu, S.; Zhu, H.; Zhang, S.; Wang, R.; Ning, C.; et al. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Vet. Res. 2017, 13, 158. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Xiao, L.; Ma, J.; Guo, M.; Liu, L.; Feng, Y. Anthroponotic enteric parasites in monkeys in public park, China. Emerg. Infect. Dis. 2012, 18, 1640–1643. [Google Scholar] [CrossRef] [PubMed]
- Karim, M.R.; Dong, H.; Li, T.; Yu, F.; Li, D.; Zhang, L.; Li, J.; Wang, R.; Li, S.; Li, X.; et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: High genetic diversity and zoonotic significance. PLoS ONE 2015, 10, e0117991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Li, W.; Zhong, Z.; Gong, C.; Cao, X.; Song, Y.; Wang, W.; Huang, X.; Liu, X.; Hu, Y.; et al. Multi-locus genotypes of Enterocytozoon bieneusi in captive Asiatic black bears in southwestern China: High genetic diversity, broad host range, and zoonotic potential. PLoS ONE 2017, 12, e0171772. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.H.; Du, S.Z.; Wang, H.B.; Hu, X.F.; Deng, M.J.; Yu, S.K.; Zhang, L.X.; Zhu, X.Q. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect. Genet. Evol. 2015, 34, 394–401. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, K.; Zhang, Y.; Wang, K.; Gazizova, A.; Wang, L.; Cao, L.; Zhang, Y.; Huang, J.; Cui, Y.; et al. First detection of Enterocytozoon bieneusi in whooper swans (Cygnus cygnus) in China. Parasites Vectors 2020, 13, 5. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Li, W.; Liu, H.; Zhong, Z.; Luo, Y.; Wei, Y.; Fu, W.; Ren, Z.; Zhou, Z.; Deng, L.; et al. First report of Giardia duodenalis and Enterocytozoon bieneusi in forest musk deer (Moschus berezovskii) in China. Parasites Vectors 2018, 11, 204. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Tian, Y.; Song, Y.; Deng, L.; Li, J.; Ren, Z.; Ma, X.; Gu, X.; He, C.; Geng, Y.; et al. Molecular characterization and multi-locus genotypes of Enterocytozoon bieneusi from captive red kangaroos (Macropus Rfus) in Jiangsu province, China. PLoS ONE 2017, 12, e0190660. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Yue, C.J.; Chai, Y.J.; Wang, W.Y.; Su, X.Y.; Zhou, Z.Y.; Wang, L.Q.; Li, L.Y.; Liu, H.F.; Zhong, Z.J.; et al. New genotypes and molecular characterization of Enterocytozoon bieneusi in pet birds in Southwestern China. Int. J. Parasitol. Parasites Wildl. 2019, 10, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhong, Z.; Song, Y.; Gong, C.; Deng, L.; Cao, Y.; Zhou, Z.; Cao, X.; Tian, Y.; Li, H.; et al. Human-Pathogenic Enterocytozoon bieneusi in Captive Giant Pandas (Ailuropoda melanoleuca) in China. Sci. Rep. 2018, 8, 6590. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Zhou, Z.; Liu, H.; Deng, L.; Bi, B.; Chai, Y.; Zhong, Z.; Hu, Y.; Fu, H.; Peng, G. New genotypes and molecular characterization of Enterocytozoon bieneusi in captive black bears in China. Int. J. Parasitol. Parasites Wildl. 2019, 10, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Breton, J.; Bart-Delabesse, E.; Biligui, S.; Carbone, A.; Seiller, X.; Okome-Nkoumou, M.; Nzamba, C.; Kombila, M.; Accoceberry, I.; Thellier, M. New highly divergent rRNA sequence among biodiverse genotypes of Enterocytozoon bieneusi strains isolated from humans in Gabon and Cameroon. J. Clin. Microbiol. 2007, 45, 2580–2589. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Xiao, L. Multilocus Sequence Typing and Population Genetic Analysis of Enterocytozoon bieneusi: Host Specificity and Its Impacts on Public Health. Front. Genet. 2019, 10, 307. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Zhao, X.; Zhang, L.; Zhang, G.; Guo, M.; Liu, L.; Feng, Y.; Xiao, L. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2013, 51, 557–563. [Google Scholar] [CrossRef] [Green Version]
- Cama, V.A.; Pearson, J.; Cabrera, L.; Pacheco, L.; Gilman, R.; Meyer, S.; Ortega, Y.; Xiao, L. Transmission of Enterocytozoon bieneusi between a child and guinea pigs. J. Clin. Microbiol. 2007, 45, 2708–2710. [Google Scholar] [CrossRef] [Green Version]
- Weiss, L.M.; Zhu, X.; Cali, A.; Tanowitz, H.B.; Wittner, M. Utility of microsporidian rRNA in diagnosis and phylogeny: A review. Folia Parasitol. 1994, 41, 81–90. [Google Scholar]
- Schuitema, A.; Hartskeerl, R.; Van Gool, T.; Laxminarayan, R.; Terpstra, W. Application of the polymerase chain reaction for the diagnosis of microsporidiosis. AIDS 1993, 7, S62–S63. [Google Scholar]
- Da Silva, A.J.; Slemenda, S.B.; Visvesvara, G.S.; Schwartz, D.A.; Wilcox, C.M.; Wallace, S.; Pieniazek, N.J. Detection of Septata intestinalis (Microsporidia) Cali et al. 1993 Using Polymerase Chain Reaction Primers Targeting the Small Submit Subunit Ribosomal RNA Coding Region. Mol. Diagn. 1997, 2, 47–52. [Google Scholar]
- Valenčáková, A.; Sučik, M. Alternatives in Molecular Diagnostics of Encephalitozoon and Enterocytozoon Infections. J. Fungi 2020, 6, 114. [Google Scholar] [CrossRef] [PubMed]
- Visvesvara, G.S.; Leitch, G.J.; da Silva, A.J.; Croppo, G.P.; Moura, H.; Wallace, S.; Slemenda, S.B.; Schwartz, D.A.; Moss, D.; Bryan, R.T.; et al. Polyclonal and monoclonal antibody and PCR-amplified small-subunit rRNA identification of a microsporidian, Encephalitozoon hellem, isolated from an AIDS patient with disseminated infection. J. Clin. Microbiol. 1994, 32, 2760–2768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Groote, M.A.; Visvesvara, G.; Wilson, M.L.; Pieniazek, N.J.; Slemenda, S.B.; daSilva, A.J.; Leitch, G.J.; Bryan, R.T.; Reves, R. Polymerase chain reaction and culture confirmation of disseminated Encephalitozoon cuniculi in a patient with AIDS: Successful therapy with albendazole. J. Infect. Dis. 1995, 171, 1375–1378. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Li, L.; Visvesvara, G.S.; Moura, H.; Didier, E.S.; Lal, A.A. Genotyping Encephalitozoon cuniculi by multilocus analyses of genes with repetitive sequences. J. Clin. Microbiol. 2001, 39, 2248–2253. [Google Scholar] [CrossRef] [Green Version]
- Xiao, L.; Li, L.; Moura, H.; Sulaiman, I.; Lal, A.A.; Gatti, S.; Scaglia, M.; Didier, E.S.; Visvesvara, G.S. Genotyping Encephalitozoon hellem isolates by analysis of the polar tube protein gene. J. Clin. Microbiol. 2001, 39, 2191–2196. [Google Scholar] [CrossRef] [Green Version]
- Hinney, B.; Sak, B.; Joachim, A.; Kváč, M. More than a rabbit’s tale - Encephalitozoon spp. in wild mammals and birds. Int. J. Parasitology. Parasites Wildl. 2016, 5, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Valencáková, A.; Balent, P.; Húska, M.; Novotný, F.; Luptáková, L. First report on Encephalitozoon intestinalis infection of swine in Europe. Acta Vet. Hung. 2006, 54, 407–411. [Google Scholar] [CrossRef]
- Duzlu, O.; Yildirim, A.; Onder, Z.; Ciloglu, A.; Yetismis, G.; Inci, A. Prevalence and Genotyping of Microsporidian Parasites in Dogs in Turkey: Zoonotic Concerns. J. Eukaryot. Microbiol. 2019, 66, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, S.; Tabrizi, A.S.; Bahrami, M.; Momtaz, H. Microsporidia in household dogs and cats in Iran; a zoonotic concern. Vet. Parasitol. 2012, 185, 121–123. [Google Scholar] [CrossRef]
- Jedrzejewski, S.; Graczyk, T.K.; Slodkowicz-Kowalska, A.; Tamang, L.; Majewska, A.C. Quantitative assessment of contamination of fresh food produce of various retail types by human-virulent microsporidian spores. Appl. Environ. Microbiol. 2007, 73, 4071–4073. [Google Scholar] [CrossRef] [Green Version]


| Region | Group (i/ii) | No. of Positive/No. of Samples (%) | PCR Positive | |||
|---|---|---|---|---|---|---|
| E. bieneusi | Encephalitozoon spp. | Mixed Infection | ||||
| No. (%) | Genotype (n) | No. (%) | No. (%) | |||
| CDZ | i | 2/11 (18.2) | 1 (9.1) | D | 1 (9.0) | 0 |
| WLCPGSB | i | 1/18 (5.6) | 0 (0.0) | 0 | 1 (5.6) | 0 |
| SLR | i | 3/42 (7.1) | 3 (7.1) | SCR1 | 0 | 0 |
| BFXZ | i | 1/14 (7.1) | 1 (7.1) | D | 0 | 0 |
| CDRBGPB | ii | 22/100 (22.0) | 18 (18.0) | PL2 | 4 (4.0) | 0 |
| PV | ii | 2/13 (15.4) | 1 (7.7) | SC02 | 2 (15.4) | 1 (7.7) |
| Total | 31/198 (15.7) | 24 (12.1) | D, SCR1, PL2, SC02 | 8/198 (4.0) | 1 (0.5) | |
| Sampling Sites | ITS Genotype | Multi-Locus Genotypes | ||||
|---|---|---|---|---|---|---|
| MS1 | MS3 | MS4 | MS7 | MLGs | ||
| CDRBGPB | PL2 | - | Type 1 | - | Type 1 | - |
| PL2 | - | Type 1 | - | Type 1 | - | |
| PL2 | Type 1 | Type 1 | Type 1 | Type 1 | MLG1 | |
| PL2 | Type 1 | Type 1 | - | - | - | |
| PL2 | - | - | - | Type 1 | - | |
| PL2 | Type 1 | - | Type 1 | Type 1 | - | |
| PL2 | - | Type 1 | - | - | - | |
| PL2 | - | Type 1 | - | Type 1 | - | |
| PL2 | - | Type 1 | - | Type 1 | - | |
| PL2 | Type 1 | Type 1 | Type 2 | Type 1 | MLG2 | |
| PL2 | Type 1 | - | - | Type 1 | - | |
| PL2 | Type 1 | Type 1 | Type 3 | Type 1 | MLG3 | |
| SLR | SCR1 | - | Type 1 | - | Type 1 | - |
| BFXZ | D | - | Type 2 | - | - | - |
| PV | SC02 | - | - | - | Type 1 | - |
| Family | Species | No. of Tested | No. of Positive (%) | Reference |
|---|---|---|---|---|
| Ailuridae | Red pandas (Ailurus fulgens) | 198 | 24 (12.1%) | This study |
| Cercopithecidae | Golden snub-nosed monkey (Cercopithecus kandti) | 160 | 74 (46.2%) | [33] |
| Rhesus macaque (Macaca mulatta) | 411 | 116 (28.2%) | [34] | |
| Hamadryas baboon (Papio hamadryas) | 21 | 6 (28.6%) | [35] | |
| Cynomolgus monkey (Macaca fascicularis) | 62 | 42 (67.7%) | [26] | |
| Cebidae | Squirrel monkey (Saimiri sp.) | 43 | 17 (39.5%) | [35] |
| Black-capped capuchin (Cebus apella) | 22 | 6 (27.3%) | [35] | |
| Ursidae | Giant panda (Ailuropoda melanoleuca) | 46 | 4 (8.7%) | [19] |
| Asiatic black bears (Ursus thibetanus) | 106 | 29 (27.4%) | [36] | |
| Bovidae | Golden takin (Budorcas taxicolor bedfordi) | 191 | 28 (14.7%) | [37] |
| Anatidae | Whooper swans (Cygnus cygnus) | 467 | 35 (7.5%) | [38] |
| Moschidae | Musk deer (Moschus berezovskii) | 223 | 38 (17.0%) | [39] |
| Lemuridae | Ring-tailed lemur (Lemur catta) | 45 | 11 (24.0%) | [35] |
| Hominidae | Bornean orangutan (Pongo pygmaeus) | 23 | 4 (17.4%) | [35] |
| Macropodidae | Red kangaroo (Macropus rufus) | 38 | 14 (36.8%) | [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Zeng, Y.; Li, C.; Liu, S.; Meng, W.; Zhang, W.; He, M.; Wang, L.; Zuo, Z.; Yue, C.; et al. Occurrence and Molecular Characteristics of Microsporidia in Captive Red Pandas (Ailurus fulgens) in China. Animals 2023, 13, 1864. https://doi.org/10.3390/ani13111864
Yang J, Zeng Y, Li C, Liu S, Meng W, Zhang W, He M, Wang L, Zuo Z, Yue C, et al. Occurrence and Molecular Characteristics of Microsporidia in Captive Red Pandas (Ailurus fulgens) in China. Animals. 2023; 13(11):1864. https://doi.org/10.3390/ani13111864
Chicago/Turabian StyleYang, Jinpeng, Yangyang Zeng, Caiwu Li, Songrui Liu, Wanyu Meng, Wenqing Zhang, Ming He, Liqin Wang, Zhili Zuo, Chanjuan Yue, and et al. 2023. "Occurrence and Molecular Characteristics of Microsporidia in Captive Red Pandas (Ailurus fulgens) in China" Animals 13, no. 11: 1864. https://doi.org/10.3390/ani13111864
APA StyleYang, J., Zeng, Y., Li, C., Liu, S., Meng, W., Zhang, W., He, M., Wang, L., Zuo, Z., Yue, C., Li, D., & Peng, G. (2023). Occurrence and Molecular Characteristics of Microsporidia in Captive Red Pandas (Ailurus fulgens) in China. Animals, 13(11), 1864. https://doi.org/10.3390/ani13111864





