Photoperiod Induces DNA Methylation Changes in the Melatonin Receptor 1A Gene in Ewes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. qPCR and Western Blotting
2.3. Isolation of the 5′-Flanking Region and Luciferase Reporter Vector Construction
2.4. Cell Culture, Transfection, and Core Promoter Identification via Luciferase Assay
2.5. DNA Isolation and Bisulfite Treatment
2.6. Pyrosequencing Analysis
2.7. Statistical Analysis
3. Results
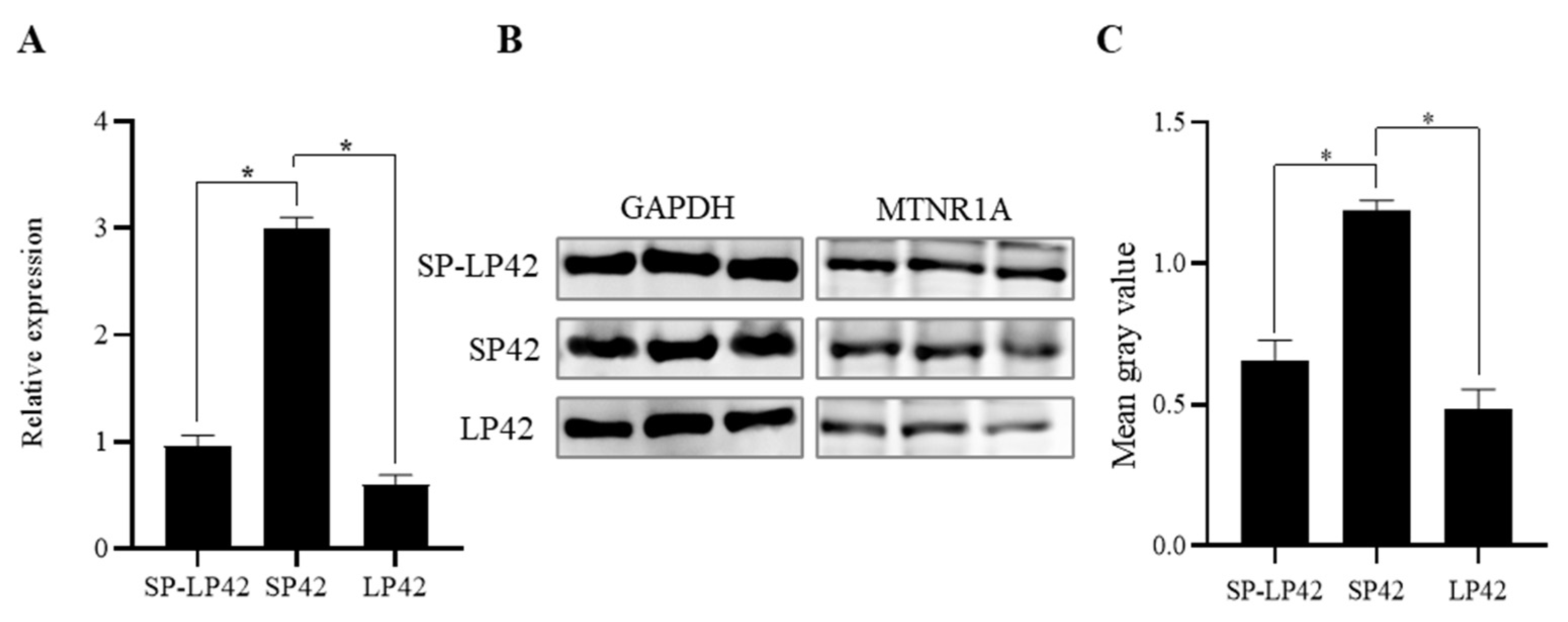
3.1. Expression Differences of MTNR1A in Different Photoperiod Groups
3.2. Identification of the Core Promoter in the MTNR1A Gene
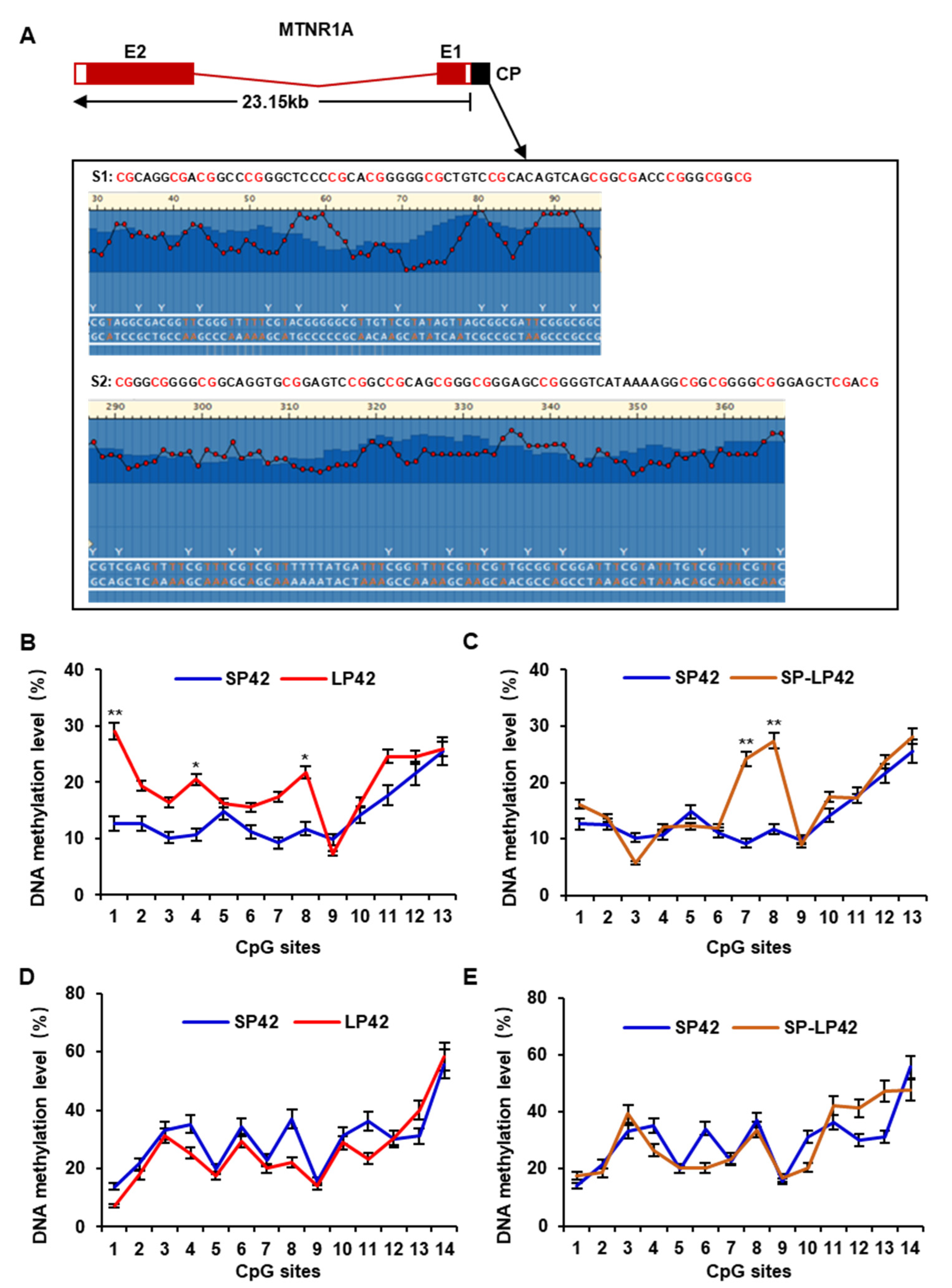
3.3. DNA Methylation Analysis of the Core Promoter in the MTNR1A Gene
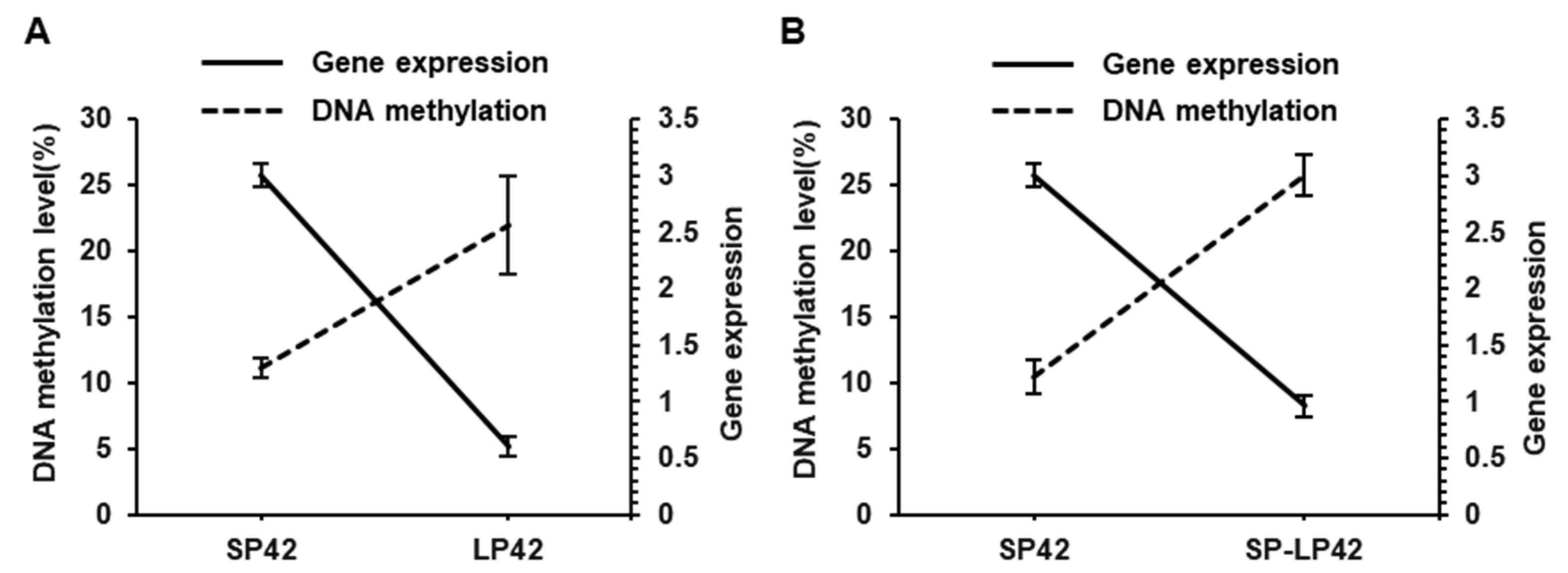
3.4. Correlation between DNA Methylation and MTNR1A Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bittman, E.L.; Karsch, F.J.; Hopkins, J.W. Role of the pineal gland in ovine photoperiodism: Regulation of seasonal breeding and negative feedback effects of estradiol upon luteinizing hormone secretion. Endocrinology 1983, 113, 329–336. [Google Scholar] [CrossRef]
- Maitra, S.K.; Hasan, K.N. The role of melatonin as a hormone and an antioxidant in the control of fish Reproduction. Front. Endocrinol. (Lausanne) 2016, 7, 38. [Google Scholar] [CrossRef]
- Sulkava, S.; Taka, A.M.; Kantojärvi, K.; Pölkki, P.; Morales-Muñoz, I.; Milani, L.; Porkka-Heiskanen, T.; Saarenpää-Heikkilä, O.; Kylliäinen, A.; Juulia, P.E.; et al. Variation near MTNR1A associates with early development and interacts with seasons. J. Sleep Res. 2020, 29, e12925. [Google Scholar] [CrossRef]
- Cosso, G.; Nehme, M.; Luridiana, S.; Pulinas, L.; Curone, G.; Hosri, C.; Carcangiu, V.; Mura, M.C. Detection of Polymorphisms in the MTNR1A gene and their association with reproductive performance in Awassi ewes. Animals 2021, 11, 583. [Google Scholar] [CrossRef]
- Zhang, J.; Jiang, H.; Du, K.; Xie, T.; Wang, B.; Chen, C.; Reiter, R.J.; Cen, B.; Yuan, Y. Pan-cancer analyses reveal genomics and clinical characteristics of the melatonergic regulators in cancer. J. Pineal Res. 2021, 71, e12758. [Google Scholar] [CrossRef]
- Zetouni, L.; de Camargo, G.M.; Da, S.F.P.; Cardoso, D.F.; Gil, F.M.; Hurtado-Lugo, N.A.; Aspilcueta-Borquis, R.R.; Cervini, M.; Tonhati, H. Polymorphisms in the MTRN1A gene and their effects on the productive and reproductive traits in buffaloes. Trop. Anim. Health Prod. 2014, 46, 337–340. [Google Scholar] [CrossRef]
- Gerdin, M.J.; Masana, M.I.; Rivera-Bermúdez, M.A.; Hudson, R.L.; Earnest, D.J.; Gillette, M.U.; Dubocovich, M.L. Melatonin desensitizes endogenous MT2 melatonin receptors in the rat suprachiasmatic nucleus: Relevance for defining the periods of sensitivity of the mammalian circadian clock to melatonin. FASEB J. 2004, 18, 1646–1656. [Google Scholar] [CrossRef]
- Martínez-Royo, A.; Lahoz, B.; Alabart, J.L.; Folch, J.; Calvo, J.H. Characterisation of the melatonin receptor 1A (MTNR1A) gene in the Rasa Aragonesa sheep breed: Association with reproductive seasonality. Anim. Reprod. Sci. 2012, 133, 169–175. [Google Scholar] [CrossRef]
- Mateescu, R.G.; Lunsford, A.K.; Thonney, M.L. Association between melatonin receptor 1A gene polymorphism and reproductive performance in Dorset ewes. J. Anim. Sci. 2009, 87, 2485–2488. [Google Scholar] [CrossRef] [PubMed]
- Luridiana, S.; Cosso, G.; Pulinas, L.; Di Stefano, M.V.; Curone, G.; Carcangiu, V.; Mura, M.C. New Polymorphisms at MTNR1A gene and their association with reproductive resumption in Sarda breed sheep. Theriogenology 2020, 158, 438–444. [Google Scholar] [CrossRef]
- Yang, D.; He, Y.; Yuan, L.; Ge, W.; Chen, S.; Chen, W.; Zhou, K.; Bao, Y. Expression and distribution of MTNR1A in the hypothalamus-pituitary-gonadal axis of Tibetan sheep during the estrous cycle. Gene 2022, 839, 146731. [Google Scholar] [CrossRef]
- He, X.Y.; Di, R.; Guo, X.F.; Cao, X.H.; Zhou, M.; Li, X.Y.; Xia, Q.; Wang, X.Y.; Zhang, J.L.; Zhang, X.S.; et al. Transcriptomic changes of photoperiodic response in the hypothalamus were identified in ovariectomized and estradiol-treated sheep. Front Mol. Biosci. 2022, 9, 848144. [Google Scholar] [CrossRef]
- Tomikawa, J.; Uenoyama, Y.; Ozawa, M.; Fukanuma, T.; Takase, K.; Goto, T.; Abe, H.; Ieda, N.; Minabe, S.; Deura, C.; et al. Epigenetic regulation of Kiss1 gene expression mediating estrogen-positive feedback action in the mouse brain. Proc. Natl. Acad. Sci. USA 2012, 109, E1294–E1301. [Google Scholar] [CrossRef]
- He, X.; Liu, Q.; Li, X.; Guo, X.; Wang, X.; Hu, W.; Di, R.; Chu, M. Molecular cloning and epigenetic change detection of Kiss1 during seasonal reproduction in Chinese indigenous sheep. Reprod Fertil. Dev. 2018, 30, 734–743. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Amodei, R.; Vilain, E.; Roselli, C.E. Identification of differential hypothalamic DNA methylation and gene expression associated with sexual partner preferences in rams. PLoS ONE 2022, 17, e0263319. [Google Scholar] [CrossRef]
- Yang, C.; Ye, J.; Li, X.; Gao, X.; Zhang, K.; Luo, L.; Ding, J.; Zhang, Y.; Li, Y.; Cao, H.; et al. DNA methylation patterns in the hypothalamus of female pubertal goats. PLoS ONE 2016, 11, e165327. [Google Scholar] [CrossRef]
- Chai, Z.; Wu, Z.; Ji, Q.; Wang, J.; Wang, J.; Wang, H.; Zhang, C.; Zhong, J.; Xin, J. Genome-wide DNA methylation and hydroxymethylation changes revealed epigenetic regulation of neuromodulation and myelination in yak hypothalamus. Front. Genet. 2021, 12, 592135. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, S.; Zhao, X.; Li, H.; Sun, J. Characterization of genome-wide DNA methylation and hydroxymethylation in mouse arcuate nucleus of hypothalamus during puberty process. Front. Genet. 2020, 11, 626536. [Google Scholar] [CrossRef]
- Stevenson, T.J.; Prendergast, B.J. Reversible DNA methylation regulates seasonal photoperiodic time measurement. Proc. Natl. Acad. Sci. USA 2013, 110, 16651–16656. [Google Scholar] [CrossRef]
- Pegoraro, M.; Bafna, A.; Davies, N.J.; Shuker, D.M.; Tauber, E. DNA methylation changes induced by long and short photoperiods in Nasonia. Genome Res. 2016, 26, 203–210. [Google Scholar] [CrossRef]
- He, X.; Tao, L.; Zhong, Y.; Di, R.; Xia, Q.; Wang, X.; Guo, X.; Gan, S.; Zhang, X.; Zhang, J.; et al. Photoperiod induced the pituitary differential regulation of lncRNAs and mRNAs related to reproduction in sheep. Peer J. 2021, 9, e10953. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, Z.; Liu, Q.; Chu, M. Polymorphisms of the melatonin receptor 1A gene that affects the reproductive seasonality and litter size in Small Tail Han sheep. Reprod. Domest. Anim. 2019, 54, 1400–1410. [Google Scholar] [CrossRef] [PubMed]
- Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br. J. Pharmacol. 2018, 175, 3190–3199. [Google Scholar] [CrossRef] [PubMed]
- Brzezinski, A.; Rai, S.; Purohit, A. Pandi-Perumal, S.R. Melatonin, clock genes, and mammalian reproduction: What is the link? Int. J. Mol. Sci. 2021, 22, 13240. [Google Scholar] [CrossRef]
- Pulinas, L.; Starič, J.; Cosso, G.; Curone, G.; Mura, M.C.; Carcangiu, V.; Luridiana, S. MTNR1A Gene Polymorphisms and reproductive recovery after seasonal anoestrus in different Mediterranean sheep breeds. Anim. Reprod. Sci. 2022, 236, 106905. [Google Scholar] [CrossRef]
- Gunwant, P.; Pandey, A.K.; Kumar, A.; Singh, I.; Kumar, S.; Phogat, J.B.; Kumar, V.; Patil, C.S.; Tomar, P.; Kumar, S.; et al. Polymorphism of melatonin receptor (MTNR1A) gene and its association with seasonal reproduction in water buffalo (Bubalus Bubalis). Anim. Reprod. Sci. 2018, 199, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Oblak, L.; van der Zaag, J.; Higgins-Chen, A.T.; Levine, M.E.; Boks, M.P. A Systematic review of biological, social and environmental factors associated with epigenetic clock acceleration. Ageing Res. Rev. 2021, 69, 101348. [Google Scholar] [CrossRef] [PubMed]
- Morris, B.J.; Willcox, B.J.; Donlon, T.A. Genetic and epigenetic regulation of human aging and longevity. Biochim. Biophys. Acta Mol. Basis. Dis. 2019, 1865, 1718–1744. [Google Scholar] [CrossRef]
- Calvo, J.H.; Serrano, M.; Martinez-Royo, A.; Lahoz, B.; Sarto, P.; Ibañez-Deler, A.; Folch, J.; Alabart, J.L. SNP Rs403212791 in exon 2 of the MTNR1A gene is associated with reproductive seasonality in the Rasa Aragonesa sheep breed. Theriogenology 2018, 113, 63–72. [Google Scholar]
- Abecia, J.A.; Mura, M.C.; Carvajal-Serna, M.; Pulinas, L.; Macías, A.; Casao, A.; Pérez-Pe, R.; Carcangiu, V. Polymorphisms of the melatonin receptor 1A (MTNR1A) gene influence the age at first mating in autumn-born ram-lambs and sexual activity of adult rams in Spring. Theriogenology 2020, 157, 42–47. [Google Scholar] [CrossRef]
- Arjoune, A.; Alsaleh, A.B.; Messaoudi, S.A.; Chelbi, H.; Jelassi, R.; Assidi, M.; Najar, T.; Haddad, B.; Sirard, M.A. Analysis of MTNR1A genetic polymorphisms and their association with the reproductive performance parameters in two Mediterranean sheep breeds. Animals 2023, 13, 448. [Google Scholar] [CrossRef]
- Starič, J.; Farci, F.; Luridiana, S.; Mura, M.C.; Pulinas, L.; Cosso, G.; Carcangiu, V. performance in three slovenian sheep breeds with different alleles for the MTNR1A gene. Anim. Reprod. Sci. 2020, 216, 106352. [Google Scholar] [CrossRef] [PubMed]
- Migaud, M.; Daveau, A.; Malpaux, B. MTNR1A Melatonin receptors in the ovine premammillary hypothalamus: Day-night variation in the expression of the transcripts. Biol. Reprod. 2005, 72, 393–398. [Google Scholar] [CrossRef] [PubMed]
- de Mendoza, A.; Nguyen, T.V.; Ford, E.; Poppe, D.; Buckberry, S.; Pflueger, J.; Grimmer, M.R.; Stolzenburg, S.; Bogdanovic, O.; Oshlack, A.; et al. Large-scale manipulation of promoter DNA methylation reveals context-specific transcriptional responses and stability. Genome Biol. 2022, 23, 163. [Google Scholar] [CrossRef]
- Alvarado, S.; Fernald, R.D.; Storey, K.B.; Szyf, M. The Dynamic nature of DNA methylation: A role in response to social and seasonal variation. Integr. Comp. Biol. 2014, 54, 68–76. [Google Scholar] [CrossRef]
- Kresovich, J.K.; Harmon, Q.E.; Xu, Z.; Nichols, H.B.; Sandler, D.P.; Taylor, J.A. Reproduction, DNA methylation and biological age. Hum. Reprod. 2019, 34, 1965–1973. [Google Scholar] [CrossRef] [PubMed]
- Takaki, N.; Uchiwa, T.; Furuse, M.; Yasuo, S. Effect of postnatal photoperiod on DNA methylation dynamics in the mouse brain. Brain Res. 2020, 1733, 146725. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacol 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA Methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Ritonja, J.A.; Aronson, K.J.; Leung, M.; Flaten, L.; Topouza, D.G.; Duan, Q.L.; Durocher, F.; Tranmer, J.E.; Bhatti, P. Investigating the relationship between melatonin patterns and methylation in circadian genes among day shift and night shift workers. Occup. Environ. Med. 2022, 79, 673–680. [Google Scholar] [CrossRef]
- Attwood, J.T.; Yung, R.L.; Richardson, B.C. DNA methylation and the regulation of gene transcription. Cell Mol. Life Sci. 2002, 59, 241–257. [Google Scholar] [CrossRef]

| Primer Name | Sequences (5′-3′) | Product Size (bp) |
|---|---|---|
| MTNR1A-F | CCTCAGATACGGCAAGCTG | 127 |
| MTNR1A-R | GATCCTCGGGTCATACTGCA | |
| ACTB-F | GCTGTATTCCCCTCCATCGT | 97 |
| ACTB-R | GGATACCTCTCTTGCTCTGG |
| Primer Name | Sequences (5′-3′) | Product Size (bp) |
|---|---|---|
| MTNR1A-F1 | AAGAAGGAGTAGGGTGTTTTTG | 274 bp |
| MTNR1A-R1 | ACTACCCTTACCCTTAAAAATCCC | |
| MTNR1A-S1 | CCCCCCCCCAAACACCTAA | |
| MTNR1A-F2 | ATGTTTATTAAGATGGTGAAGATGAG | 396 bp |
| MTNR1A-R2 | TTAAAAAAAACCCAAAATACCCTTAAA | |
| MTNR1A-S2 | GGTGGATTTTTAGAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Wang, W.; Sun, W.; Chu, M. Photoperiod Induces DNA Methylation Changes in the Melatonin Receptor 1A Gene in Ewes. Animals 2023, 13, 1917. https://doi.org/10.3390/ani13121917
He X, Wang W, Sun W, Chu M. Photoperiod Induces DNA Methylation Changes in the Melatonin Receptor 1A Gene in Ewes. Animals. 2023; 13(12):1917. https://doi.org/10.3390/ani13121917
Chicago/Turabian StyleHe, Xiaoyun, Wei Wang, Wei Sun, and Mingxing Chu. 2023. "Photoperiod Induces DNA Methylation Changes in the Melatonin Receptor 1A Gene in Ewes" Animals 13, no. 12: 1917. https://doi.org/10.3390/ani13121917






