Automatic Penaeus Monodon Larvae Counting via Equal Keypoint Regression with Smartphones
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.1.1. The Collection of Raw Larvae Data
2.1.2. The Penaeus_1k Dataset
2.2. Methods
2.2.1. Ground Truth Generation
2.2.2. Feature Extracting Module
2.2.3. Penaeus Larvae Counting Strategy
| Algorithm 1 Penaeus Larvae Counting Strategy |
| Input: predicted heatmap H generated by backbone. Output: coordinates C and quantity Q of the larvae in the input. /* {Boolean ? A:B} means returning A if it was true, otherwise B */ /* get all candidate points */
|
2.2.4. Experimental Setup
2.2.5. Method Performance Evaluation
2.3. A Smartphone App for Shrimp Larvae Counting
3. Results
3.1. Model Training Results
3.2. Image Counting Results
3.3. Comparisons with the Crowd Counting Methods
3.4. Ablative Analysis
3.5. On BBBC041v1 Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FAO. The State of World Fisheries and Aquaculture 2022; FAO: Rome, Italy, 2022. [Google Scholar]
- MARA. China Fishery Statistical Yearbook; China Agriculture Press: Beijing, China, 2022. [Google Scholar]
- Motoh, H. Biology and Ecology of Penaeus Monodon. In Proceedings of the First International Conference on the Culture of Penaeid Prawns/Shrimps, Iloilo, Philippines, 4–7 December 1984; pp. 27–36. [Google Scholar]
- Kesvarakul, R.; Chianrabutra, C.; Chianrabutra, S. Baby Shrimp Counting via Automated Image Processing. In Proceedings of the 9th International Conference on Machine Learning and Computing, Singapore, Singapore, 24–26 February 2017; pp. 352–356. [Google Scholar]
- Yada, S.; Chen, H. Weighing Type Counting System for Seedling Fry. Nippon Suisan Gakkaishi 1997, 63, 178–183. [Google Scholar] [CrossRef] [Green Version]
- Khantuwan, W.; Khiripet, N. Live Shrimp Larvae Counting Method Using Co-Occurrence Color Histogram. In Proceedings of the 2012 9th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology, Phetchaburi, Thailand, 16–18 May 2012; pp. 1–4. [Google Scholar]
- Solahudin, M.; Slamet, W.; Dwi, A. Vaname (Litopenaeus Vannamei) Shrimp Fry Counting Based on Image Processing Method. IOP Conf. Ser. Earth Environ. Sci. 2018, 147, 012014. [Google Scholar] [CrossRef]
- Kaewchote, J.; Janyong, S.; Limprasert, W. Image Recognition Method Using Local Binary Pattern and the Random Forest Classifier to Count Post Larvae Shrimp. Agric. Nat. Resour. 2018, 52, 371–376. [Google Scholar] [CrossRef]
- Yeh, C.-T.; Chen, M.-C. A Combination of IoT and Cloud Application for Automatic Shrimp Counting. Microsyst. Technol. 2022, 28, 187–194. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, X.; Li, B.; Zhang, H.; Duan, Q. Automatic Shrimp Counting Method Using Local Images and Lightweight YOLOv4. Biosyst. Eng. 2022, 220, 39–54. [Google Scholar] [CrossRef]
- Lainez, S.M.D.; Gonzales, D.B. Automated Fingerlings Counting Using Convolutional Neural Network. In Proceedings of the 2019 IEEE 4th International Conference on Computer and Communication Systems, Singapore, 23–25 February 2019; pp. 67–72. [Google Scholar]
- Nguyen, K.-T.; Nguyen, C.-N.; Wang, C.-Y.; Wang, J.-C. Two-Phase Instance Segmentation for Whiteleg Shrimp Larvae Counting. In Proceedings of the 2020 IEEE International Conference on Consumer Electronics, Las Vegas, NV, USA, 4–6 January 2020; pp. 1–3. [Google Scholar]
- Hong Khai, T.; Abdullah, S.N.H.S.; Hasan, M.K.; Tarmizi, A. Underwater Fish Detection and Counting Using Mask Regional Convolutional Neural Network. Water 2022, 14, 222. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. Yolov3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. Yolov4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Wang, C.-Y.; Bochkovskiy, A.; Liao, H.-Y.M. YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. Yolox: Exceeding Yolo Series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. Adv. Neural Inf. Process. Syst. 2015, 28, 91–99. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhou, D.; Chen, S.; Gao, S.; Ma, Y. Single-Image Crowd Counting via Multi-Column Convolutional Neural Network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 589–597. [Google Scholar]
- Li, Y.; Zhang, X.; Chen, D. CSRNet: Dilated Convolutional Neural Networks for Understanding the Highly Congested Scenes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1091–1100. [Google Scholar]
- Ma, Z.; Wei, X.; Hong, X.; Gong, Y. Bayesian Loss for Crowd Count Estimation With Point Supervision. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 6142–6151. [Google Scholar]
- Tian, Y.; Chu, X.; Wang, H. Cctrans: Simplifying and Improving Crowd Counting with Transformer. arXiv 2021, arXiv:2109.14483. [Google Scholar]
- Fan, S.; Lin, X.; Zhou, P. Research on Automatic Counting of Shrimp Fry Based on Improved Convolutional Neural Network. Fish. Mod. 2020, 47, 35. [Google Scholar]
- Liu, W.; Salzmann, M.; Fua, P. Context-Aware Crowd Counting. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 5099–5108. [Google Scholar]
- Wang, Q.; Meng, J. A Shrimp Seedling Density Estimation Method Based on Improved Unet. Mod. Inf. Technol. 2021, 5, 12–16. [Google Scholar]
- Gao, J.; Han, T.; Wang, Q.; Yuan, Y.; Li, X. Learning Independent Instance Maps for Crowd Localization. arXiv 2020, arXiv:2012.04164. [Google Scholar]
- Wang, J.; Sun, K.; Cheng, T.; Jiang, B.; Deng, C.; Zhao, Y.; Liu, D.; Mu, Y.; Tan, M.; Wang, X.; et al. Deep High-Resolution Representation Learning for Visual Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2020, 43, 3349–3364. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-Y.; Liao, H.-Y.M.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W.; Yeh, I.-H. CSPNet: A New Backbone That Can Enhance Learning Capability of CNN. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, Seattle, WA, USA, 13–19 June 2020; pp. 390–391. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Ljosa, V.; Sokolnicki, K.L.; Carpenter, A.E. Annotated High-Throughput Microscopy Image Sets for Validation. Nat. Methods 2012, 9, 637. [Google Scholar] [CrossRef] [Green Version]
- Rahman, A.; Zunair, H.; Reme, T.R.; Rahman, M.S.; Mahdy, M.R.C. A Comparative Analysis of Deep Learning Architectures on High Variation Malaria Parasite Classification Dataset. Tissue Cell 2021, 69, 101473. [Google Scholar] [CrossRef]
- Doering, E.; Pukropski, A.; Krumnack, U.; Schaffand, A. Automatic Detection and Counting of Malaria Parasite-Infected Blood Cells. In Proceedings of the Medical Imaging and Computer-Aided Diagnosis, Singapore, 3 July 2020; pp. 145–157. [Google Scholar]
- Depto, D.S.; Rahman, S.; Hosen, M.M.; Akter, M.S.; Reme, T.R.; Rahman, A.; Zunair, H.; Rahman, M.S.; Mahdy, M. Automatic Segmentation of Blood Cells from Microscopic Slides: A Comparative Analysis. Tissue Cell 2021, 73, 101653. [Google Scholar]
- Manku, R.R.; Sharma, A.; Panchbhai, A. Malaria Detection and Classificaiton. arXiv 2020, arXiv:2011.14329. [Google Scholar]
- Rocha, M.; Claro, M.; Neto, L.; Aires, K.; Machado, V.; Veras, R. Malaria Parasites Detection and Identification Using Object Detectors Based on Deep Neural Networks: A Wide Comparative Analysis. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2022, 11, 1–18. [Google Scholar]



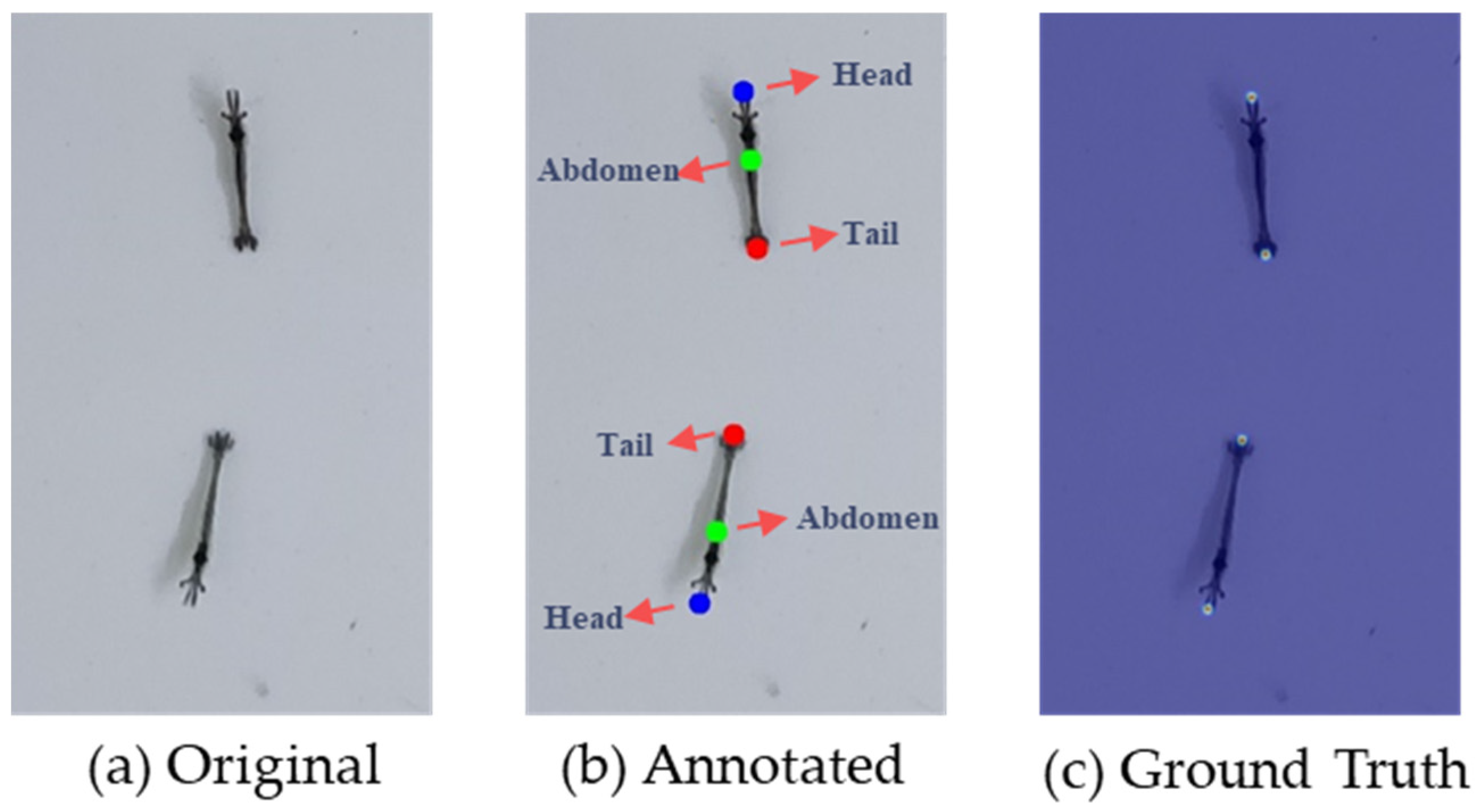

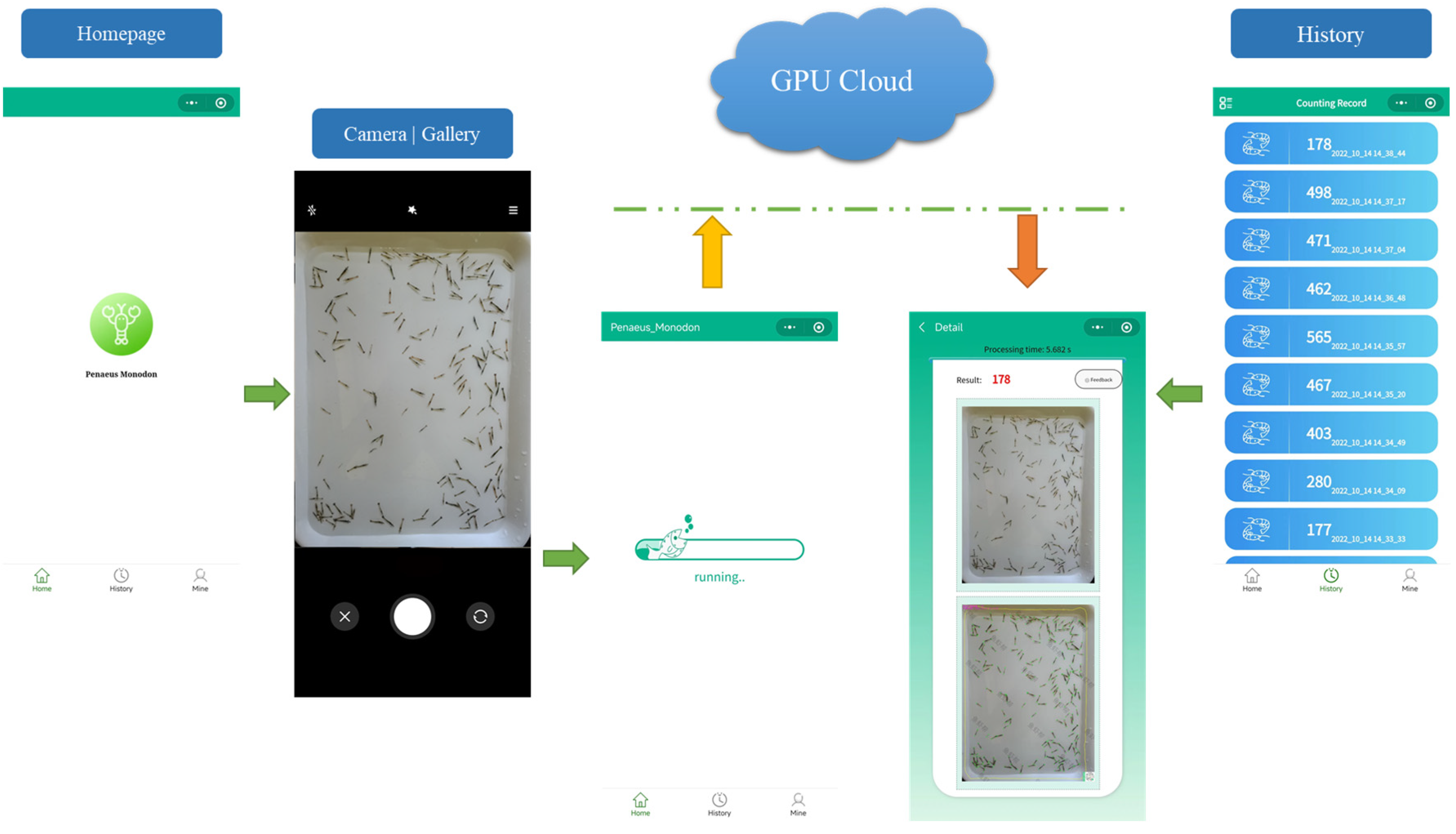


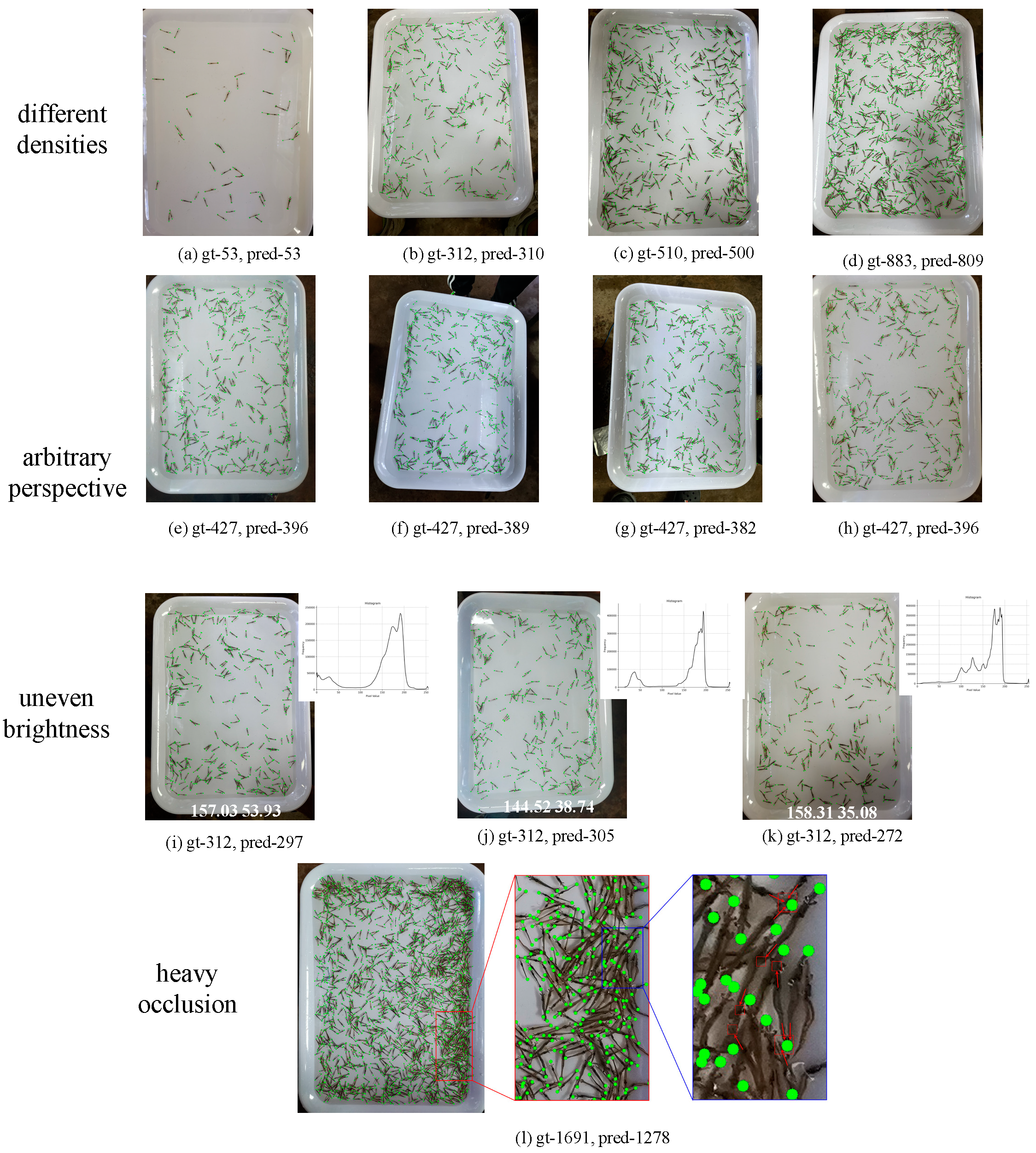



| Device Model | Main Cameras | Image Resolution | Shooting Mode |
|---|---|---|---|
| iPhone 11 | 12 MP (wide), 12 MP (ultrawide) | 4032 × 3024 | Auto |
| iPhone 13 | 12 MP (wide), 12 MP (ultrawide) | 4032 × 3024 | Auto |
| Redmi K40 | 48 MP (wide), 8 MP (ultrawide), 5 MP (macro) | 3456 × 4608 | Auto |
| Huawei P20 | 12 MP(wide), 20 MP(wide) | 2736 × 3648 | Auto |
| Huawei P50 Pro | 50 MP (wide), 64 MP (periscope telephoto), 13 MP (ultrawide), 40 MP (B/W) | 3072 × 4096 | Auto |
| Group | The Number of Larvae |
|---|---|
| 1 | 53 |
| 2 | 183 |
| 3 | 312 |
| 4 | 427 |
| 5 | 510 |
| 6 | 675 |
| 7 | 883 |
| 8 | 1691 |
| Parameter | Value |
|---|---|
| Epoch | 50 |
| σ | 3 |
| Batch size | 4 |
| Input size | 1024 |
| Heatmap size | 256 |
| Optimizer | Adam |
| Learning rate | 0.0015 |
| Metric | Result |
|---|---|
| Acc | 93.79% |
| MAE | 33.69 |
| MSE | 34.74 |
| Method | Accuracy (%) | MAE | MSE |
|---|---|---|---|
| CSRNet | 63.94 | 105.00 | 126.61 |
| BLNet | 82.42 | 52.18 | 62.75 |
| CCTrans | 86.13 | 47.42 | 58.33 |
| Ours | 93.79 | 33.69 | 45.30 |
| Group | Keypoints | Accuracy (%) | MAE | MSE |
|---|---|---|---|---|
| 1 | head, abdomen, tail | 77.95 | 104.28 | 108.59 |
| 2 | head, tail | 93.79 | 33.69 | 34.74 |
| 3 | head, abdomen | 77.73 | 102.69 | 105.53 |
| 4 | abdomen, tail | 85.88 | 47.56 | 53.13 |
| 5 | head | 83.44 | 77.91 | 80.50 |
| 6 | abdomen | 90.87 | 49.11 | 50.76 |
| 7 | tail | 84.99 | 77.18 | 79.78 |
| Method | Acc (%) | MAE | MSE |
|---|---|---|---|
| BLNet | 80.87 | 8.53 | 11.11 |
| CCTrans | 82.21 | 8.33 | 11.74 |
| Ours-w48 | 82.33 | 7.49 | 8.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Liu, R.; Wang, Z.; Zheng, G.; Lv, J.; Fan, L.; Guo, Y.; Gao, Y. Automatic Penaeus Monodon Larvae Counting via Equal Keypoint Regression with Smartphones. Animals 2023, 13, 2036. https://doi.org/10.3390/ani13122036
Li X, Liu R, Wang Z, Zheng G, Lv J, Fan L, Guo Y, Gao Y. Automatic Penaeus Monodon Larvae Counting via Equal Keypoint Regression with Smartphones. Animals. 2023; 13(12):2036. https://doi.org/10.3390/ani13122036
Chicago/Turabian StyleLi, Ximing, Ruixiang Liu, Zhe Wang, Guotai Zheng, Junlin Lv, Lanfen Fan, Yubin Guo, and Yuefang Gao. 2023. "Automatic Penaeus Monodon Larvae Counting via Equal Keypoint Regression with Smartphones" Animals 13, no. 12: 2036. https://doi.org/10.3390/ani13122036






