Goose Hepatic IGFBP2 Is Regulated by Nutritional Status and Participates in Energy Metabolism Mainly through the Cytokine−Cytokine Receptor Pathway
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Sample Collection
2.2. Isolation and Culture of Goose Primary Hepatocytes
2.3. Glucose and Insulin Treatments of Goose Primary Hepatocytes
2.4. Overexpression and Interference Assays of IGFBP2 Gene in Goose Primary Hepatocytes
2.5. Total RNA Isolation and qRT-PCR Analysis
2.6. Transcriptome Analysis
2.7. Statistical Analysis
3. Results
3.1. Effect of Changes in Nutritional Status on IGFBP2 Expression in Goose Liver
3.2. Effect of Glucose and Insulin on IGFBP2 Expression in Goose Primary Hepatocytes
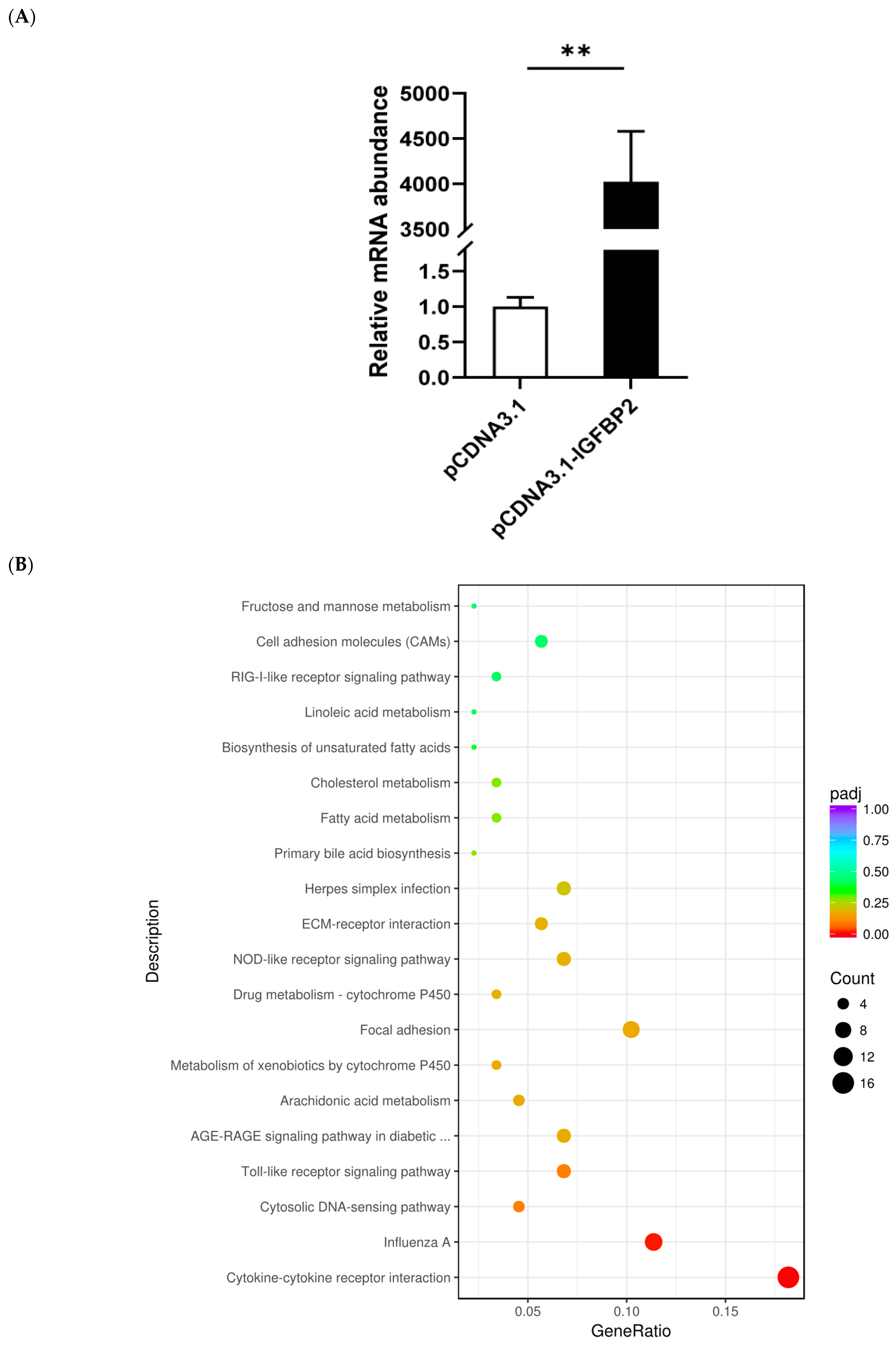
3.3. Downstream Genes and Pathways Affected by IGFBP2 Overexpression
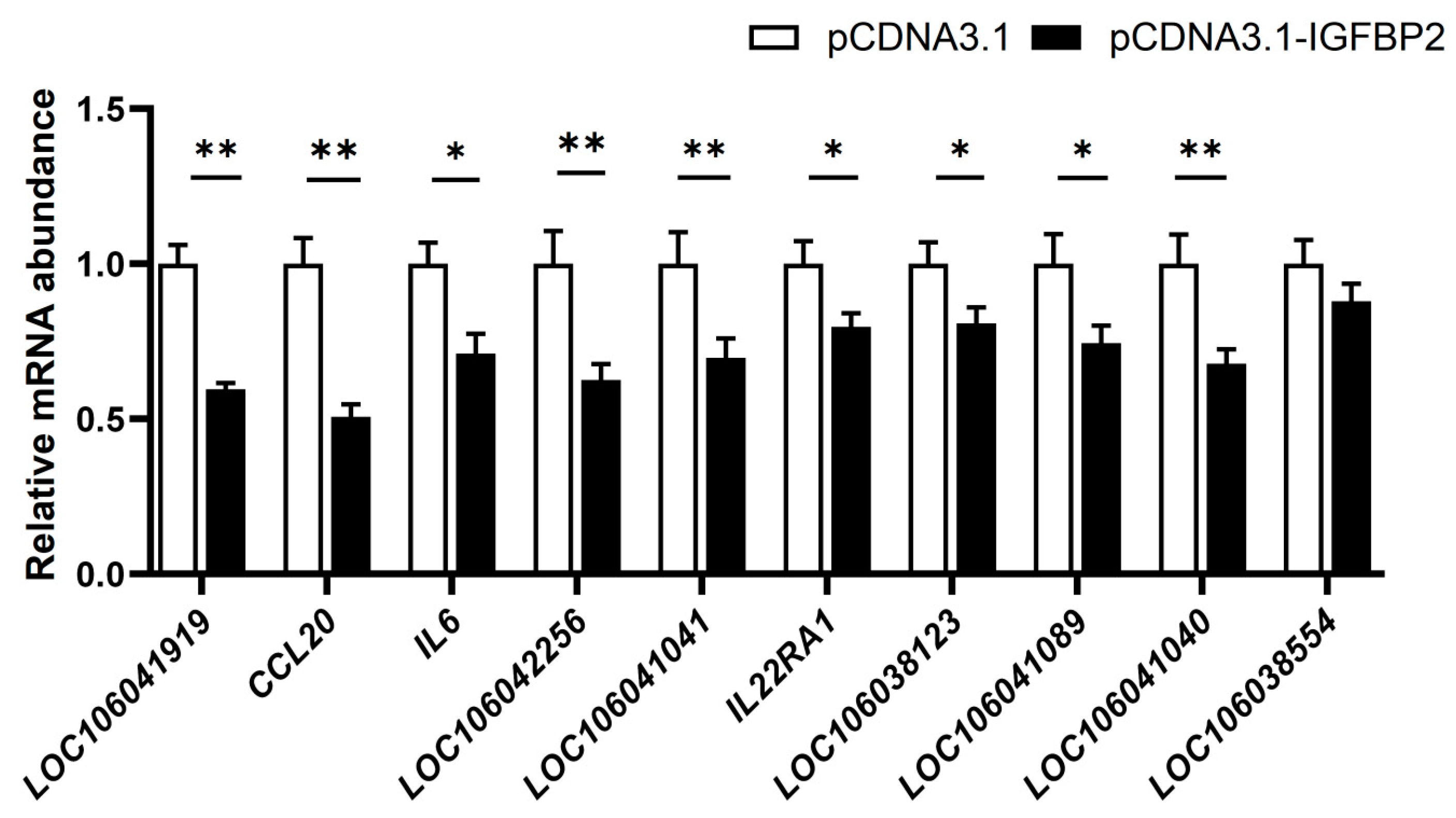
3.4. Effect of IGFBP2 Overexpression on the Expression of the Genes in the Cytokine−Cytokine Receptor Pathway
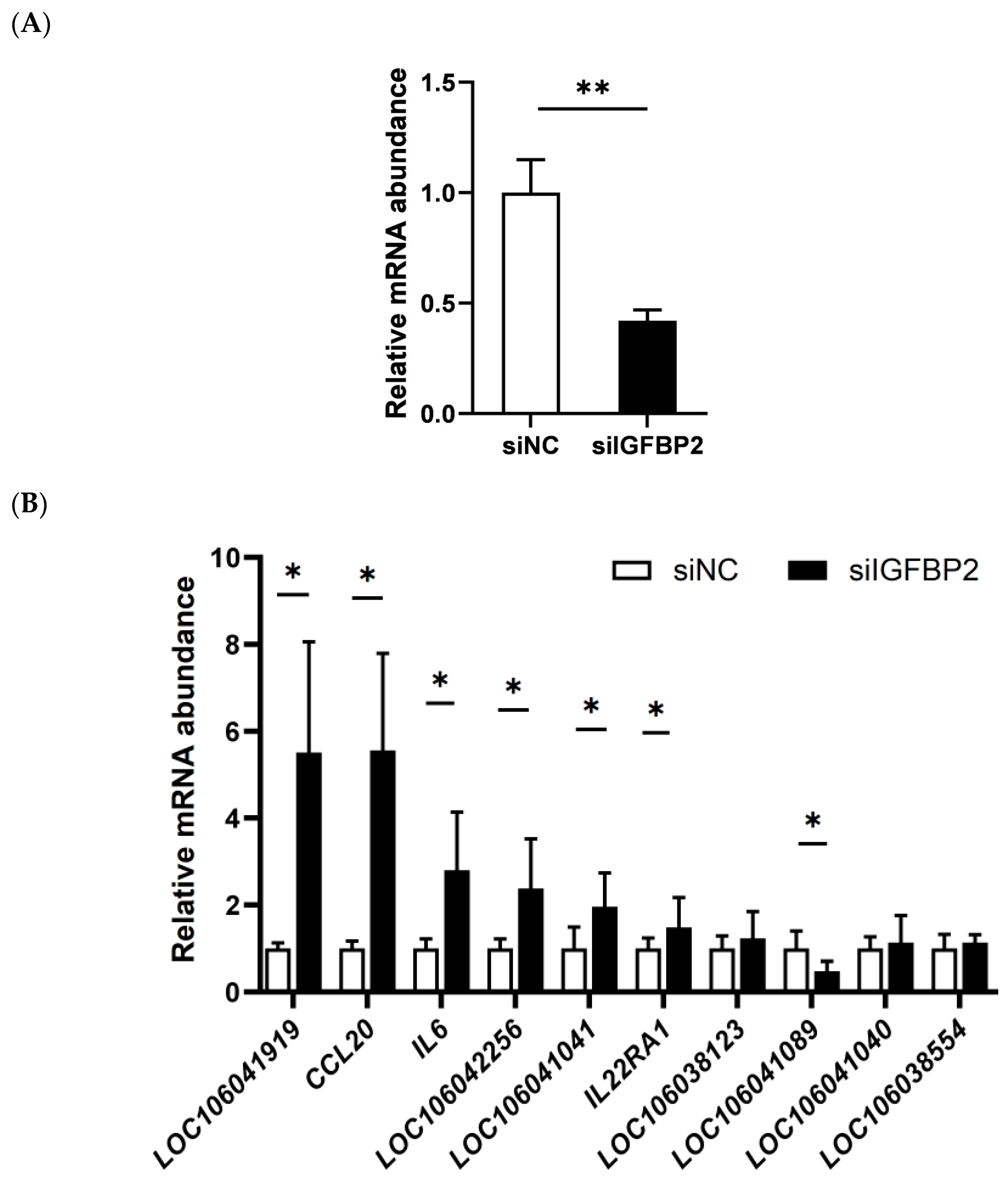
3.5. Effect of IGFBP2 Interference on the Expression of the Genes in the Cytokine−Cytokine Receptor Pathway
3.6. Effect of Overfeeding on the Expression of the Genes in the Cytokine−Cytokine Receptor Pathway in Goose Liver
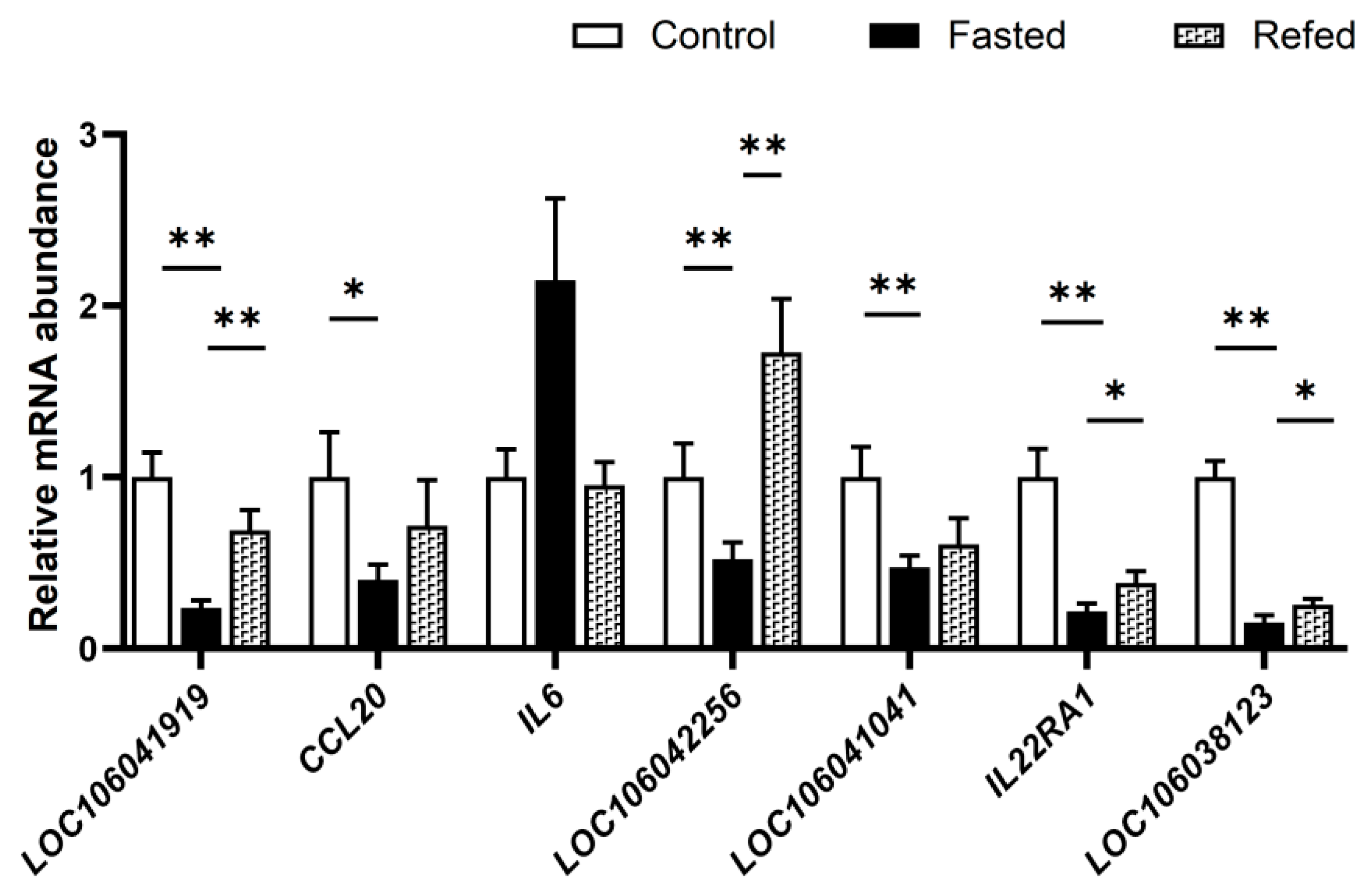
3.7. Effect of Fasting and Refeeding on the Expression of the Genes in the Cytokine−Cytokine Receptor Pathway in Goose Liver
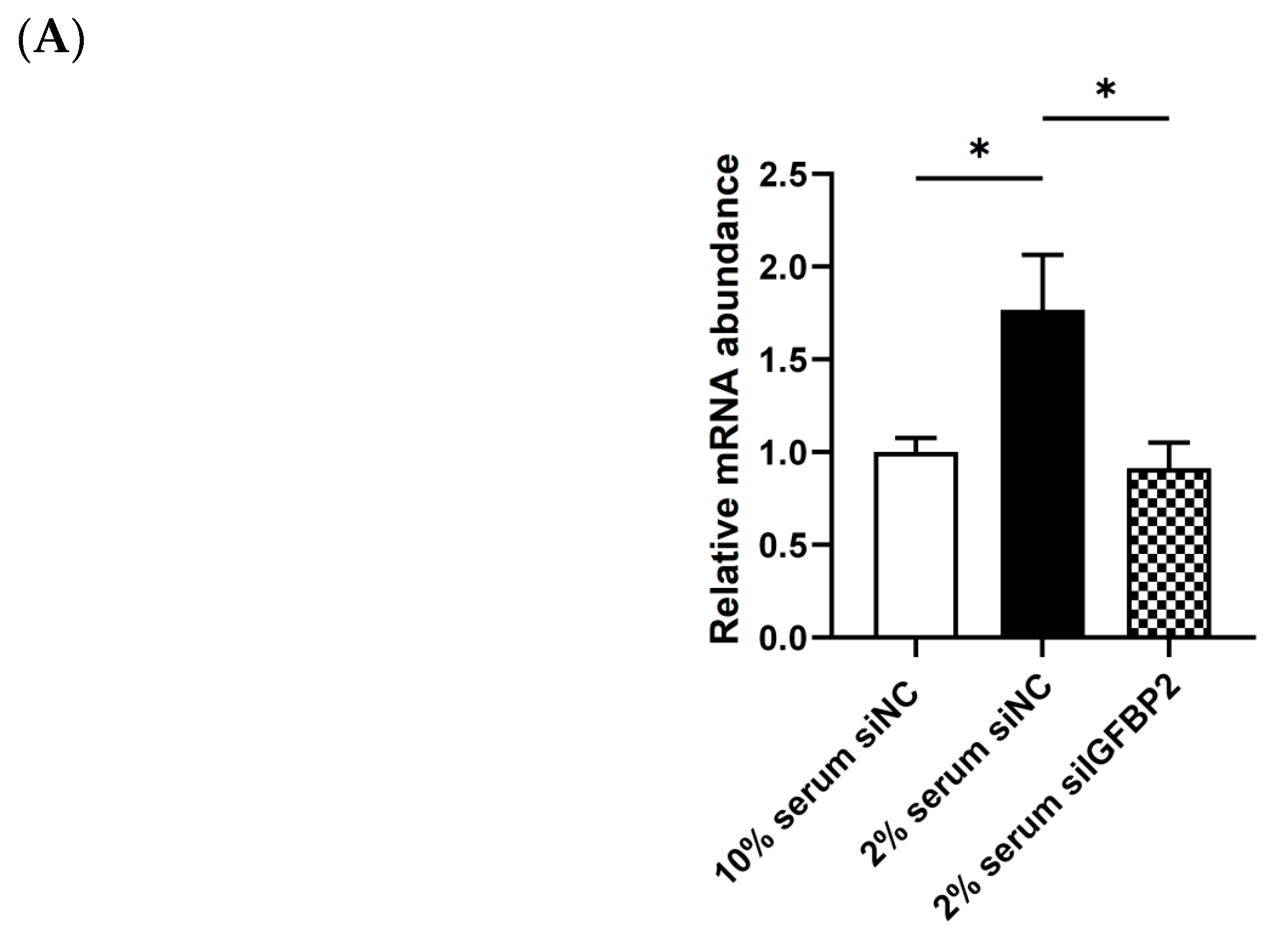
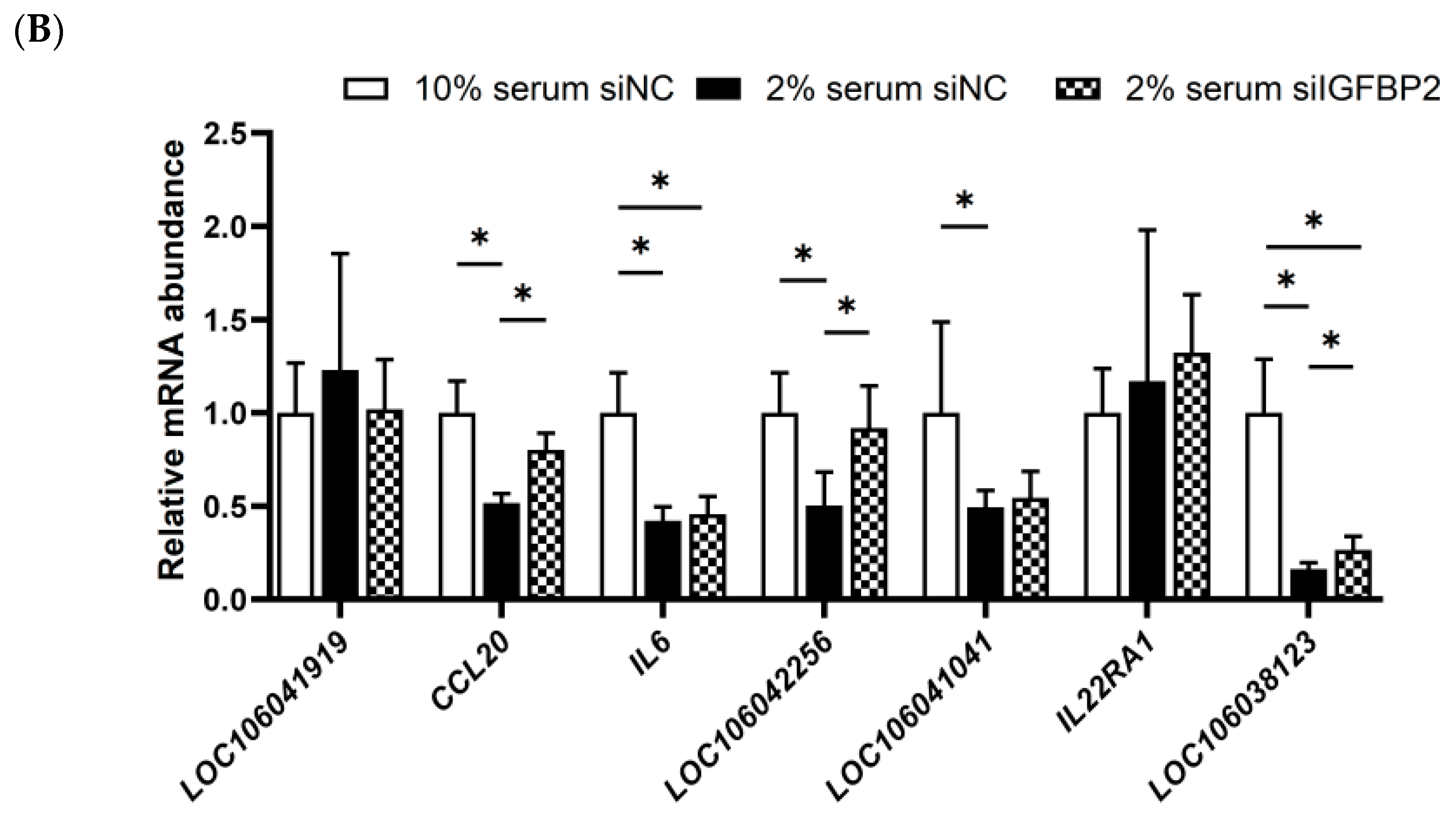
3.8. Validation of IGFBP2 Regulating the Expression of the Genes in the Cytokine−Cytokine Receptor Pathway
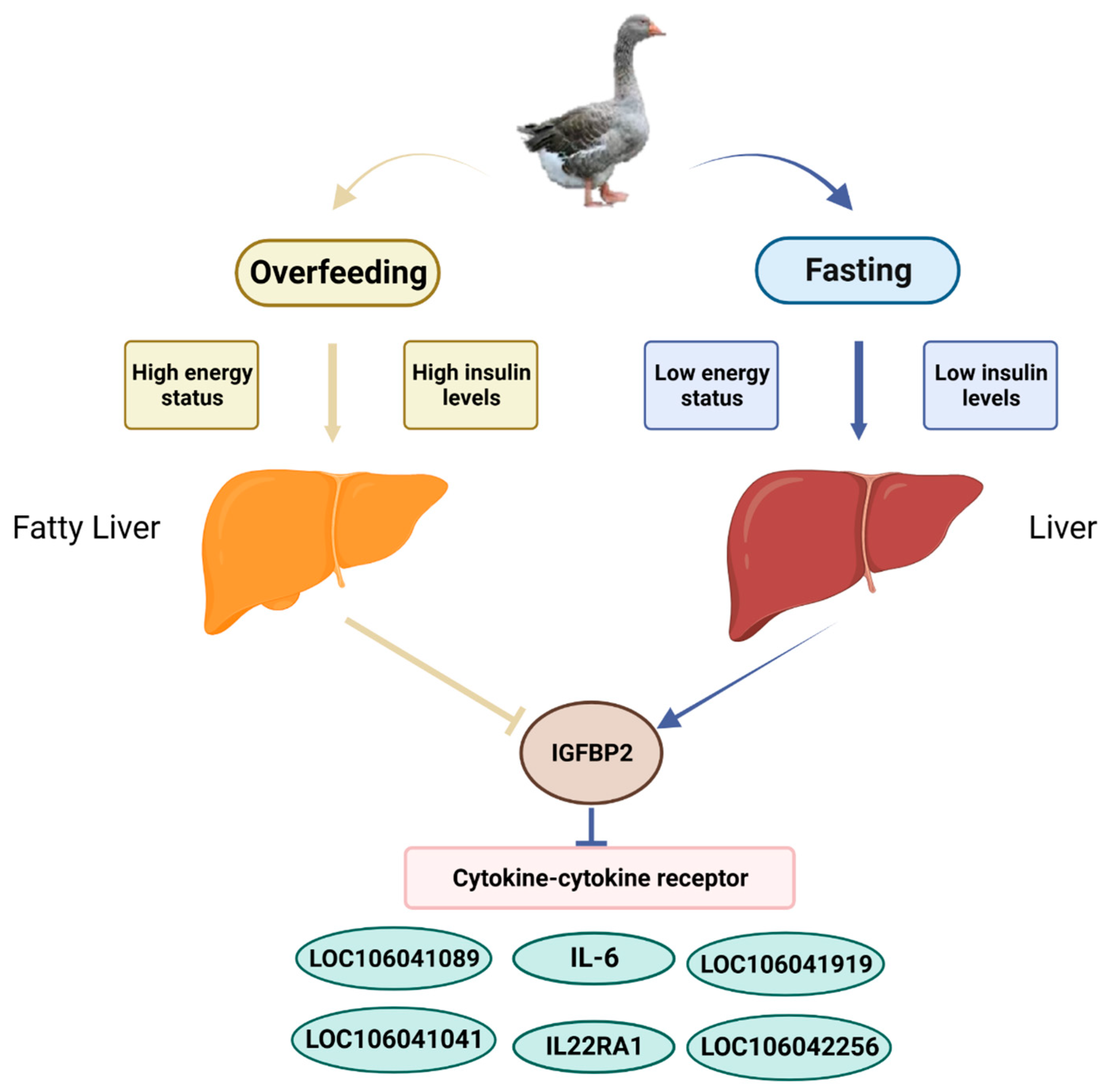
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, L.; Peng, X.; Han, F.; Wu, F.; Chen, D.; Wu, D.; Feyera, T.; Zhang, K.; Che, L. Effects of birth weight and postnatal nutritional restriction on skeletal muscle development, myofiber maturation, and metabolic status of early-weaned piglets. Animals 2020, 10, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Hu, H.; Xu, Y.; Wang, D.; Jiang, L.; Li, K.; Wang, Y.; Zhan, X. Effects of posthatch feed deprivation on residual yolk absorption, macronutrients synthesis, and organ development in broiler chicks. Poult. Sci. 2020, 99, 5587–5597. [Google Scholar] [CrossRef]
- Parsons, A.S.; Buchanan, N.P.; Blemings, K.P.; Wilson, M.E.; Moritz, J.S. Effect of corn particle size and pellet texture on broiler performance in the growing phase. J. Appl. Poult. Res 2006, 15, 245–255. [Google Scholar] [CrossRef]
- Bobyleva-Guarriero, V.; Hughes, P.E.; Ronchetti-Pasquali, I.; Lardy, H.A. The influence of fasting on chicken liver metabolites, enzymes and mitochondrial respiration. Comp. Biochem. Physiol. Part B 1984, 78, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Reyns, G.E.; Janssens, K.A.; Buyse, J.; Kuhn, E.R.; Darras, V.M. Changes in thyroid hormone levels in chicken liver during fasting and refeeding. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2002, 132, 239–245. [Google Scholar] [CrossRef]
- Simon, J.; Rosebrough, R.W.; McMurtry, J.P.; Steele, N.C.; Roth, J.; Adamo, M.; LeRoith, D. Fasting and refeeding alter the insulin receptor tyrosine kinase in chicken liver but fail to affect brain insulin receptors. J. Biol. Chem. 1986, 261, 17081–17088. [Google Scholar] [CrossRef]
- Xiang, L.; Jiao, Y.; Qian, Y.; Li, Y.; Mao, F.; Lu, Y. Comparison of hepatic gene expression profiles between three mouse models of nonalcoholic gatty liver disease. Genes Dis. 2022, 9, 201–215. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, X.; Wang, Q.; Sun, X.; Xia, L.; Wang, Q.; Yang, B.; Zhang, Y.; Montgomery, S.; Meng, H. Prosteatotic and protective components in a unique model of fatty liver: Gut microbiota and suppressed complement system. Sci. Rep. 2016, 6, 31763. [Google Scholar] [CrossRef] [Green Version]
- Pliszka, M.; Szablewski, L.; Oleszczak, B. Effects of IGF-1 on deexy-D-glucose uptake by human peripheral blood lymphocytes. Diabetol. Dosw. Klin. 2006, 6, 314–319. [Google Scholar]
- Fuentes, E.N.; Bjornsson, B.T.; Valdes, J.A.; Einarsdottir, I.E.; Lorca, B.; Alvarez, M.; Molina, A. IGF-I/PI3K/Akt and IGF-I/MAPK/ERK pathways in vivo in skeletal muscle are regulated by nutrition and contribute to somatic growth in the fine flounder. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2011, 300, 1532–1542. [Google Scholar] [CrossRef]
- Houston, B.; O’Neill, I.E. Insulin and growth hormone act synergistically to stimulate insulin-like growth factor-I production by cultured chicken hepatocytes. J. Endocrinol. 1991, 128, 389–393. [Google Scholar] [CrossRef]
- Xi, G.; Solum, M.A.; Wai, C.; Maile, L.A.; Rosen, C.J.; Clemmons, D.R. The heparin-binding domains of IGFBP-2 mediate its inhibitory effect on preadipocyte differentiation and fat development in male mice. Endocrinology 2013, 154, 4146–4157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoen, T.J.; Mazuruk, K.; Waldbillig, R.J.; Potts, J.; Beebe, D.C.; Chader, G.J.; Rodriguez, I.R. Cloning and characterization of a chick embryo cDNA and gene for IGF-binding protein-2. J. Mol. Endocrnol. 1995, 15, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Fan, Z.; Liu, D.; Wang, F.; Zhou, Y. IGFBP2 enhances adipogenic differentiation potentials of mesenchymal stem cells from Wharton’s jelly of the umbilical cord via JNK and Akt signaling pathways. PLoS ONE 2017, 12, e018418. [Google Scholar] [CrossRef] [Green Version]
- Hoeflich, A.; Wu, M.; Mohan, S.; Föll, J.; Wanke, R.; Froehlich, T.; Arnold, G.J.; Lahm, H.; Kolb, H.J.; Wolf, E. Overexpression of insulin-like growth factor-binding protein-2 in transgenic mice reduces postnatal body weight gain. Endocrinology 1999, 140, 5488–5496. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.H.; Li, H.; Zhang, H.; Wang, S.Z.; Wang, Q.G.; Wang, Y.X. Identification of a single nucleotide polymorphism of the insulin-like growth factor binding protein 2 gene and its association with growth and body composition traits in the chicken. J. Anim. Sci. 2006, 84, 2902–2906. [Google Scholar] [CrossRef] [Green Version]
- Mu, Y.; Sun, Y.; Leng, L.; Cao, Z.; Li, Y.; Li, H.; Wang, Y. Effect of IGFBP2 overexpression on the expression of fatty acid synthesis genes in primary cultured chicken hepatocytes. J. Poult. Sci. 2019, 56, 177–185. [Google Scholar] [CrossRef]
- Tsukada, A. Thyroid hormones are involved in insulin-like growth factor (IGF)-I production by regulating growth hormone receptor (GHR) in the chicken. Growth Horm. IGF Res. 1998, 8, 235–242. [Google Scholar] [CrossRef]
- Ahmadi, M.R.; Kadivar, A.; Gheisari, H.R. Serum IGF-I, fatty acids, beta hydroxybutyrate and glucose before and after parturition in cows with endometritis and ovarian cysts. OJVR 2021, 25, 807–814. [Google Scholar]
- Kita, K.; Nagao, K.; Taneda, N.; Inagaki, Y.; Hirano, K.; Shibata, T.; Yaman, M.A.; Conlon, M.A.; Okumura, J.I. Insulin-Like growth factor binding protein-2 gene expression can be regulated by diet manipulation in several tissues of young chickens1. J. Nutr. 2002, 132, 145–151. [Google Scholar] [CrossRef] [Green Version]
- Kammel, A.; Saussenthaler, S.; Jahnert, M.; Jonas, W.; Stirm, L.; Hoeflich, A.; Staiger, H.; Fritsche, A.; Haring, H.U.; Joost, H.G.; et al. Early hypermethylation of hepatic Igfbp2 results in its reduced expression preceding fatty liver in mice. Hum. Mol. Genet. 2016, 25, 2588–2599. [Google Scholar]
- Hedbacker, K.; Birsoy, K.; Wysocki, R.W.; Asilmaz, E.; Ahima, R.S.; Farooqi, I.S.; Friedman, J.M. Antidiabetic effects of IGFBP2, a leptin-regulated gene. Cell Metab. 2010, 11, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osman, R.H.; Shao, D.; Liu, L.; Xia, L.L.; Sun, X.X.; Zheng, Y.; Wang, L.D.; Zhang, R.; Zhang, Y.H.; Zhang, J.; et al. Expression of mitochondria-related genes is elevated in overfeeding-induced goose fatty liver. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2016, 192, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Geng, T.; Xia, L.; Li, F.; Xia, J.; Zhang, Y.; Wang, Q.; Yang, B.; Montgomery, S.; Cui, H.; Gong, D. The role of endoplasmic reticulum stress and insulin resistance in the occurrence of goose fatty liver. Biochem. Biophys. Res. Commun. 2015, 465, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Zhao, M.; Wen, K.; Fan, X.; Sun, Q.; Jauregui, D.; Khogali, M.; Liu, L.; Geng, T.; Gong, D. OTUD7A regulates inflammation- and immune-related gene expression in goose fatty liver. Agriculture 2022, 12, 105. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Asadi, N.B.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59–74. [Google Scholar] [CrossRef] [Green Version]
- Desert, C.; Duclos, M.J.; Blavy, P.; Lecerf, F.; Moreews, F.; Klopp, C.; Aubry, M.; Herault, F.; Le Roy, P.; Berri, C.; et al. Transcriptome profiling of the feeding-to-fasting transition in chicken liver. BMC Genomics. 2008, 9, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Hicks, J.A.; Porter, T.E.; Sunny, N.E.; Liu, H.C. Delayed feeding alters transcriptional and post-transcriptional regulation of hepatic metabolic pathways in peri-hatch broiler chicks. Genes 2019, 10, 272. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Xing, Y.; Fan, X.; Liu, T.; Zhao, M.; Liu, L.; Hu, X.; Cui, H.; Geng, T.; Gong, D. Fasting and refeeding affect the goose liver transcriptome mainly through the PPAR signaling pathway. J. Poult. Sci. 2021, 58, 245–257. [Google Scholar] [CrossRef]
- Cogburn, L.A.; Trakooljul, N.; Wang, X.; Ellestad, L.E.; Porter, T.E. Transcriptome analyses of liver in newly-hatched chicks during the metabolic perturbation of fasting and re-feeding reveals THRSPA as the key lipogenic transcription factor. BMC Genom. 2020, 21, 109. [Google Scholar] [CrossRef]
- Ma, X.; Bai, Y. IGF-1 activates the P13K/AKT signaling pathway via upregulation of secretory clusterin. Mol. Med. Rep. 2012, 6, 1433–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, A.C.; Vaillancourt, R.R. Tyrosine kinase receptor-activated signal transduction pathways which lead to oncogenesis. Oncogene 1998, 17, 1343–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Yang, G. IL22RA1/JAK/STAT signaling acts as a cancer target through pan-cancer analysis. Front. Immunol. 2022, 13, 915246. [Google Scholar] [CrossRef] [PubMed]
- Adachi, Y.; Aoki, C.; Yoshio-Hoshino, N.; Takayama, K.; Curiel, D.T.; Nishimoto, N. Interleukin-6 induces both cell growth and VEGF production in malignant mesotheliomas. Int. J. Cancer 2006, 119, 1303–1311. [Google Scholar] [CrossRef]
- Polachi, N.; Bai, G.; Li, T.; Chu, Y.; Wang, X.; Li, S.; Gu, N.; Wu, J.; Li, W.; Zhang, Y.; et al. Modulatory effects of silibinin in various cell signaling pathways against liver disorders and cancer—A comprehensive review. Eur. J. Med. Chem. 2016, 123, 577–595. [Google Scholar] [CrossRef]
- Nandi, B.; Del Valle, J.P.; Samur, M.K.; Gibbons, A.J.; Prabhala, R.H.; Munshi, N.C.; Gold, J.S. CCL20 induces colorectal cancer neoplastic epithelial cell proliferation, migration, and further CCL20 production through autocrine HGF-c-Met and MSP-MSPR signaling pathways. Oncotarget 2021, 12, 2323–2337. [Google Scholar] [CrossRef]
- Danielpour, D.; Song, K. Cross-talk between IGF-I and TGF-β signaling pathways. Cytokine Growth Factor Rev. 2006, 17, 59–74. [Google Scholar] [CrossRef]
- Jia, G.; Mitra, A.K.; Gangahar, D.M.; Agrawal, D.K. Insulin-like growth factor-1 induces phosphorylation of PI3K-Akt/PKB to potentiate proliferation of smooth muscle cells in human saphenous vein. Exp. Mol. Pathol. 2010, 89, 20–26. [Google Scholar] [CrossRef] [Green Version]
- Uehara, T.; Tokumitsu, Y.; Nomura, Y. Pertussis toxin-sensitive and insensitive intracellular signalling pathways in undifferentiated 3T3-L1 cells stimulated by insulin converge with phosphatidylinositol 3-kinase upstream of the Ras mitogen-activated protein kinase cascade. Eur. J. Biochem. 1999, 259, 801–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandhu, M.S.; Dunger, D.B.; Giovannucci, E.L. Insulin, insulin-like growth factor-I (IGF-I), IGF binding proteins, their biologic interactions, and colorectal cancer. J. Natl. Cancer Inst. 2002, 94, 972–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Q.; Dai, E.; Chen, M.; Zhang, J.; Mu, J.; Liu, L.; Geng, T.; Gong, D.; Zhang, Y.; Zhao, M. Glucose-induced enhanced anti-oxidant activity inhibits apoptosis in goose fatty liver. J. Anim. Sci. 2023, 101, skad059. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Xing, Y.; Fan, X.; Chen, Z.; Geng, T. Fasting and overfeeding affect the expression of the immunity- or inflammation-related genes in the liver of poultry via endogenous retrovirus (ERV). Poult. Sci. 2020, 100, 973–981. [Google Scholar] [CrossRef]
- Tanaka, K.; Sen, T.; Shigeno, K. The effect of fastig and refeeding on lipids of serum and liver in the meat-type chicken. Nihon Chikusan Gakkaiho 1975, 46, 396–402. [Google Scholar] [CrossRef]




| Gene Name | Gene Symbol | Log2 (Fold Change) | p-Value | Adjusted p-Value |
|---|---|---|---|---|
| Up-regulated | ||||
| Radical S-Adenosyl Methionine Domain Containing 2 | RSAD2 | 0.93 | 1.07 × 10−31 | 3.01 × 10−28 |
| Solute Carrier Family 40 Member 1 | SLC40A1 | 0.85 | 4.03 × 10−19 | 3.24 × 10−16 |
| Sorbin And SH3 Domain Containing 2 | SORBS2 | 1.14 | 4.56 × 10−13 | 1.77 × 10−10 |
| Fibrinogen Beta Chain | FGB | 0.57 | 1.95 × 10−12 | 6.67 × 10−10 |
| Lecithin-Cholesterol Acyltransferase | LCAT | 0.69 | 2.64 × 10−12 | 8.49 × 10−10 |
| Monoacylglycerol O-Acyltransferase 1 | MOGAT1 | 0.56 | 7.49 × 10−11 | 1.72 × 10−8 |
| Fibrinogen Gamma Chain | FGG | 0.47 | 4.67 × 10−10 | 9.56 × 10−8 |
| Collectin Subfamily Member 10 | COLEC10 | 1.34 | 7.46 × 10−10 | 1.50 × 10−7 |
| Adenomatosis Polyposis Coli Down-Regulated 1 Protein | APCDD1 | 0.53 | 4.33 × 10−9 | 7.74 × 10−7 |
| D-Dopachrome Tautomerase | LOC106031299 | 0.48 | 7.94 × 10−9 | 1.35 × 10−6 |
| Down-regulated | ||||
| Uncharacterized LOC106034664 | LOC106034664 | −1.65 | 1.56 × 10−43 | 8.79 × 10−40 |
| C-C Motif Chemokine 5-like | LOC106041040 | −1.36 | 2.44 × 10−38 | 9.17 × 10−35 |
| Cystatin-Like | LOC106044772 | −1.05 | 3.99 × 10−29 | 7.50 × 10−26 |
| Toll-like Receptor 2 | LOC106042256 | −2.07 | 4.86 × 10−28 | 7.81 × 10−25 |
| Extracellular Fatty Acid-binding Protein-like | LOC106037025 | −1.01 | 1.15 × 10−25 | 1.62 × 10−22 |
| Coactosin Like F-Actin Binding Protein 1 | COTL1 | −0.70 | 1.11 × 10−24 | 1.39 × 10−21 |
| Solute Carrier Family 13 Member 5 | SLC13A5 | −1.50 | 1.29 × 10−23 | 1.46 × 10−20 |
| Protein MRP-126 | LOC106049124 | −0.75 | 5.03 × 10−23 | 5.16 × 10−20 |
| Chimerin 2 | CHN2 | −0.87 | 1.60 × 10−22 | 1.50 × 10−19 |
| CD9 Molecule | CD9 | −1.05 | 1.86 × 10−20 | 1.61 × 10−17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Xing, Y.; Zhang, J.; Mu, J.; Ge, J.; Zhao, M.; Liu, L.; Gong, D.; Geng, T. Goose Hepatic IGFBP2 Is Regulated by Nutritional Status and Participates in Energy Metabolism Mainly through the Cytokine−Cytokine Receptor Pathway. Animals 2023, 13, 2336. https://doi.org/10.3390/ani13142336
Li F, Xing Y, Zhang J, Mu J, Ge J, Zhao M, Liu L, Gong D, Geng T. Goose Hepatic IGFBP2 Is Regulated by Nutritional Status and Participates in Energy Metabolism Mainly through the Cytokine−Cytokine Receptor Pathway. Animals. 2023; 13(14):2336. https://doi.org/10.3390/ani13142336
Chicago/Turabian StyleLi, Fangbo, Ya Xing, Jinqi Zhang, Ji’an Mu, Jing Ge, Minmeng Zhao, Long Liu, Daoqing Gong, and Tuoyu Geng. 2023. "Goose Hepatic IGFBP2 Is Regulated by Nutritional Status and Participates in Energy Metabolism Mainly through the Cytokine−Cytokine Receptor Pathway" Animals 13, no. 14: 2336. https://doi.org/10.3390/ani13142336
APA StyleLi, F., Xing, Y., Zhang, J., Mu, J., Ge, J., Zhao, M., Liu, L., Gong, D., & Geng, T. (2023). Goose Hepatic IGFBP2 Is Regulated by Nutritional Status and Participates in Energy Metabolism Mainly through the Cytokine−Cytokine Receptor Pathway. Animals, 13(14), 2336. https://doi.org/10.3390/ani13142336






