Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Viral Detection and Identification
2.3. CoV Characterization via Phylogenetic Analysis of Partial Sequences of the RdRp Gene
3. Results
3.1. Detection and Identification of CoV Genera and Subgenera
3.2. Phylogenetic Analysis of the RdRp Gene
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Méndez, M.P. Herencia textil, identidad indigena y recursos económicos en la Patagonia Argentina. Estudio de un caso: La comarca de La Meseta Central de La Provincia de Chubut. AIBR-Rev. Antropol. Iberoam. 2009, 4, 11–53. [Google Scholar] [CrossRef]
- Consejo Nacional de Camélidos Sudamericanos (CONACS). Estrategia Nacional de Desarrollo: Los Camélidos Domésticos en el Perú. Ministerio de Agricultura. Available online: https://www.whyalpaca.com/assets/documentos/estrategia_nacional.pdf (accessed on 31 July 2023).
- FAO (Food and Agriculture Organization of the United Nations). Situación Actual de los Camelidos Sudamericanos en el Perú. Organización de las Naciones Unidas Para la Agricultura y Alimentación. Proyecto de Cooperación Técnica en Apoyo a la Crianza y Aprovechamiento de los Camelidos Sudamericanos en la Region Andino. Available online: http://tarwi.lamolina.edu.pe/~emellisho/zootecnia_archivos/situacion%20alpcas%20peru.pdf (accessed on 31 July 2023).
- MINAGRI (Ministerio de Desarrollo Agrario y Riego/Peru). Situacion de las Actividades de Crianza y Producción de camelidos Sudamericanos en el Peru. MINAGRI. Available online: https://www.midagri.gob.pe/portal/datero/40-sector-agrario/situacion-de-las-actividades-de-crianza-y-producci/298-camelidos-sudamericanos?start=2 (accessed on 31 July 2023).
- Ramon, P.T. Trayectorias Comunales Cambios y Continuidades en Comunidades Campesinas e Indígenas del sur Andino, Segunda; GRUPO Propussta Ciudadana, Ed.; Fordfoundation: Lima, Perú, 2019. [Google Scholar]
- Pinn, T.L.; Gagliardo, L.F.; Purdy, S.R.; Appleton, J.A.; Stokol, T. Comparison of three immunoglobulin G assays for the diagnosis of failure of passive transfer of immunity in neonatal alpacas. J. Vet. Diagn. Investig. 2013, 25, 91–98. [Google Scholar] [CrossRef]
- Weaver, D.M.; Tyler, J.W.; Marion, R.S.; Wallace, L.M.; Nagy, J.K.; Holle, J.M. Evaluation of Assays for Determination of Passive Transfer Status in Neonatal Llamas and Alpacas. J. Am. Vet. Med. Assoc. 2000, 216, 559–563. [Google Scholar] [CrossRef]
- Carhuapoma Delacruz, V.; Valencia Mamani, N.; Paucar-Chanca, R.; Salas Contreras, W.H.; Morales-Cauti, S. Caracterización de neumonías y bacterias neumónicas en alpacas (Vicugna pacos) en comunidades altoandinas de Huancavelica, Perú. La. Granja 2022, 37, 75–85. [Google Scholar] [CrossRef]
- Castilla, D.; Escobar, V.; Ynga, S.; Llanco, L.; Manchego, A.; Lázaro, C.; Navarro, D.; Santos, N.; Rojas, M. Enteric viral infections among domesticated South American camelids: First detection of mammalian orthoreovirus in camelids. Animals 2021, 11, 1455. [Google Scholar] [CrossRef] [PubMed]
- Wellington López, P.; Marycris Chamorro, L.; Antonio, B. Rapid detection of rotavirus and coronavirus in alpaca crias (Vicugna pacos) with diarrhea in the Cusco region, Peru. Rev. Invest. Vet. Perú 2011, 22, 407–411. [Google Scholar] [CrossRef]
- Luna, L.; Brandão, P.E.; Maturrano, L.; Rosadio, R.; Silva, F.D.F.; Soares, R.M.; Gregori, F. Betacoronavirus 1 in alpacas (Vicugna pacos) in the High Peruvian Andes. Small Rumin. Res. 2015, 133, 7–9. [Google Scholar] [CrossRef]
- Rojas, M.A.; Gonçalves, J.L.S.; Dias, H.G.; Manchego, A.; Santos, N. Identification of two novel rotavirus A genotypes, G35 and P[50], from Peruvian alpaca faeces. Infect. Genet. Evol. 2017, 55, 71–74. [Google Scholar] [CrossRef]
- Cebra, C.K.; Mattson, D.E.; Baker, R.J.; Sonn, R.J.; Dearing, P.L. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J. Am. Vet. Med. Assoc. 2003, 223, 1806–1808. [Google Scholar] [CrossRef]
- Cirilo, C.E.; Manchego, S.A.; Rivera, H.H.; Rosadio, A.R. Coexistencia de virus y bacterias en neumonías agudas en alpacas neonatas. Rev. Invest. Vet. Perú. 2012, 23, 317–335. [Google Scholar] [CrossRef][Green Version]
- Rosadio, R.; Cirilo, E.; Manchego, A.; Rivera, H. Respiratory syncytial and parainfluenza type 3 viruses coexisting with Pasteurella multocida and Mannheimia hemolytica in acute pneumonias of neonatal alpacas. Small Rumin. Res. 2011, 97, 110–116. [Google Scholar] [CrossRef]
- Amer, H.M. Bovine-like coronaviruses in domestic and wild ruminants. Anim. Health Res. Rev. 2018, 19, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef]
- ICTV. International Committee on Taxonomy of Viruses: Coronaviridae. 2023. Available online: https://talk.ictvonline.org/taxonomy/ (accessed on 31 July 2023).
- Bashor, L.; Gagne, R.B.; Bosco-Lauth, A.M.; Bowen, R.A.; Stenglein, M.; VandeWoude, S. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl. Acad. Sci. USA 2021, 118, e2105253118. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16. [Google Scholar] [CrossRef]
- Corman, V.M.; Eckerle, I.; Memish, Z.A.; Liljander, A.M.; Dijkman, R.; Jonsdottir, H.; Juma Ngeiywa, K.J.; Kamau, E.; Younan, M.; Al Masri, M.; et al. Link of a ubiquitous human coronavirus to dromedary camels. Proc. Natl. Acad. Sci. USA 2016, 113, 9864–9869. [Google Scholar] [CrossRef] [PubMed]
- Bakkers, M.J.; Lang, Y.; Feitsma, L.J.; Hulswit, R.J.; de Poot, S.A.; van Vliet, A.L.; Margine, I.; de Groot-Mijnes, J.D.; van Kuppeveld, F.J.; Langereis, M.A.; et al. Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity. Cell. Host Microbe 2017, 21, 356–366. [Google Scholar] [CrossRef]
- Vijgen, L.; Keyaerts, E.; Lemey, P.; Maes, P.; Van Reeth, K.; Nauwynck, H.; Pensaert, M.; Van Ranst, M. Evolutionary history of the closely related group 2 coronaviruses: Porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J. Virol. 2006, 80, 7270–7274. [Google Scholar] [CrossRef]
- Falkenberg, S.; Buckley, A.; Laverack, M.; Martins, M.; Palmer, M.V.; Lager, K.; Diel, D.G. Experimental Inoculation of Young Calves with SARS-CoV-2. Viruses 2021, 13, 441. [Google Scholar] [CrossRef]
- Das, T.; Sikdar, S.; Chowdhury, M.H.U.; Nyma, K.J.; Adnan, M. SARS-CoV-2 prevalence in domestic and wildlife animals: A genomic and docking based structural comprehensive review. Heliyon 2023, 9, e19345. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Fusco, G.; Cardillo, L.; Levante, M.; Brandi, S.; Picazio, G.; Napoletano, M.; Martucciello, A.; Fiorito, F.; De Carlo, E.; de Martinis, C. First serological evidence of SARS-CoV-2 natural infection in small ruminants: Brief report. Vet. Res. Commun. 2023, 47, 1741–1748. [Google Scholar] [CrossRef]
- El Masry, I.; Al Makhladi, S.; Al Abdwany, M.; Al Subhi, A.; Eltahir, H.; Cheng, S.; Peiris, M.; Gardner, E.; Von Dobschuetz, S.; Soumare, B.; et al. Serological evidence of SARS-CoV-2 infection in dromedary camels and domestic bovids in Oman. Emerg. Microbes Infect. 2023, 12, 2220577. [Google Scholar] [CrossRef]
- Ruiz-Arrondo, I.; Portillo, A.; Palomar, A.M.; Santibáñez, S.; Santibáñez, P.; Cervera, C.; Oteo, J.A. Detection of SARS-CoV-2 in pets living with COVID-19 owners diagnosed during the COVID-19 lockdown in Spain: A case of an asymptomatic cat with SARS-CoV-2 in Europe. Transbound. Emerg. Dis. 2021, 68, 973–976. [Google Scholar] [CrossRef]
- Hosie, M.J.; Epifano, I.; Herder, V.; Orton, R.J.; Stevenson, A.; Johnson, N.; MacDonald, E.; Dunbar, D.; McDonald, M.; Howie, F.; et al. Detection of SARS-CoV-2 in respiratory samples from cats in the UK associated with human-to-cat transmission. Vet. Rec. 2021, 188, e247. [Google Scholar] [CrossRef]
- Haake, C.; Cook, S.; Pusterla, N.; Murphy, B. Coronavirus Infections in Companion Animals: Virology, Epidemiology, Clinical and Pathologic Features. Viruses 2020, 12, 1023. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Saif, L.J. Bovine Coronavirus and the Associated Diseases. Front. Vet. Sci. 2021, 8, 643220. [Google Scholar] [CrossRef]
- Savard, C.; Provost, C.; Ariel, O.; Morin, S.; Fredrickson, R.; Gagnon, C.A.; Broes, A.; Wang, L. First report and genomic characterization of a bovine-like coronavirus causing enteric infection in an odd-toed non-ruminant species (Indonesian tapir, Acrocodia indica) during an outbreak of winter dysentery in a zoo. Transbound. Emerg. Dis. 2022, 69, 3056–3065. [Google Scholar] [CrossRef]
- Erles, K.; Toomey, C.; Brooks, H.W.; Brownlie, J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. 2003, 310, 216–223. [Google Scholar] [CrossRef]
- Zhang, X.M.; Herbst, W.; Kousoulas, K.G.; Storz, J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J. Med. Virol. 1994, 44, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Rojas, M.; Manchego, A.; Rocha, C.B.; Fornells, L.A.; Silva, R.C.; Mendes, G.S.; Dias, H.G.; Sandoval, N.; Pezo, D.; Santos, N. Outbreak of diarrhea among preweaning alpacas (Vicugna pacos) in the Southern Peruvian Highland. J. Infect. Dev. Ctries. 2016, 10, 269–274. [Google Scholar] [CrossRef]
- Rocha, C.B.; Fornells, L.A.A.M.G.; Rojas, M.; Libetal, M.; Manchego, A.; Pezo, D.; Santos, N. Molecular epidemiology of coronavirus in faeces of Brazilian calves and Peruvian camelid herds. J. Infect. Dev. Ctries. 2018, 12, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Cebra, C.K.; Baker, R.J.; Mattson, D.E.; Cohen, S.A.; Alvarado, D.E.; Rohrmann, G.F. Analysis of the genome sequence of an alpaca coronavirus. Virology 2007, 365, 198–203. [Google Scholar] [CrossRef]
- Crossley, B.M.; Mock, R.E.; Callison, S.A.; Hietala, S.K. Identification and characterization of a novel alpaca respiratory coronavirus most closely related to the human coronavirus 229E. Viruses 2012, 4, 3689–3700. [Google Scholar] [CrossRef] [PubMed]
- Moës, E.; Vijgen, L.; Keyaerts, E.; Zlateva, K.; Li, S.; Maes, P.; Pyrc, K.; Berkhout, B.; van der Hoek, L.; Van Ranst, M. A novel pancoronavirus RT-PCR assay: Frequent detection of human coronavirus NL63 in children hospitalized with respiratory tract infections in Belgium. BMC Infect. Dis. 2005, 5, 6. [Google Scholar] [CrossRef]
- Brandão, P.E. Coronavírus bovino (BCoV): Ocorrência, Diversidade Molecular e Padronização de PCR Para Diagnóstico a Partir de Amostras Fecais de Bezerros com e sem Diarréia Criados em Municípios dos Estados de São Paulo e Minas Gerais, Brasil. Ph.D. Thesis, Universidade de São Paulo, São Paulo, Brazil, 2004. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Lucas, L.J.R.; Morales, C.S.; Barrios, A.M.; Rodríguez, G.J.; Vásquez, C.M.; Lira, M.B.; Torres, L.B.; Casas, A.E.; Espinoza, B.J. Patógenos involucrados en casos fatales de diarrea en crías de alpaca de la sierra central del Perú. Rev. Invest. Vet. Perú 2016, 27, 159–175. [Google Scholar] [CrossRef][Green Version]
- Martín Espada, M.; Pinto Jiménez, C.; Cid Vázquez, M. Camélidos Sudamericanos: Estado sanitario de sus crías. Rev. Complut. Cienc. Vet. (RCCV) 2010, 4, 37–50. [Google Scholar]
- Islam, A.; Ferdous, J.; Sayeed, M.A.; Islam, S.; Rahman, M.K.; Abedin, J.; Saha, O.; Hassan, M.M.; Shirin, T. Spatial epidemiology and genetic diversity of SARS-CoV-2 and related coronaviruses in domestic and wild animals. PLoS ONE 2021, 16, e0260635. [Google Scholar] [CrossRef]
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an ancestral association of human coronavirus 229E with bats. J. Virol. 2015, 89, 11858–11870. [Google Scholar] [CrossRef]
- Anthony, S.J.; Johnson, C.K.; Greig, D.J.; Kramer, S.; Che, X.; Wells, H.; Hicks, A.L.; Joly, D.O.; Wolfe, N.D.; Daszak, P.; et al. Global patterns in coronavirus diversity. Virus Evol. 2017, 3, vex012. [Google Scholar] [CrossRef]
- Bueno, L.M.; Rizotto, L.S.; Viana, A.O.; Silva, L.M.N.; de Moraes, M.V.D.S.; Benassi, J.C.; Scagion, G.P.; Dorlass, E.G.; Lopes, B.L.T.; Cunha, I.N.; et al. High genetic diversity of alphacoronaviruses in bat species (Mammalia: Chiroptera) from the atlantic forest in Brazil. Transbound. Emerg. Dis. 2022, 69, e2863–e2875. [Google Scholar] [CrossRef]
- Chidoti, V.; De Nys, H.; Pinarello, V.; Mashura, G.; Missé, D.; Guerrini, L.; Pfukenyi, D.; Cappelle, J.; Chiweshe, N.; Ayouba, A.; et al. Longitudinal survey of coronavirus circulation and diversity in insectivorous bat colonies in Zimbabwe. Viruses 2022, 14, 781. [Google Scholar] [CrossRef]
- So, R.T.Y.; Chu, D.K.W.; Miguel, E.; Perera, R.A.P.M.; Oladipo, J.O.; Fassi-Fihri, O.; Aylet, G.; Ko, R.L.W.; Zhou, Z.; Cheng, M.-S.; et al. camel coronavirus HKU23 in African camels revealed multiple recombination events among closely related Betacoronaviruses of the subgenus Embecovirus. J. Virol. 2019, 93, e01236-19. [Google Scholar] [CrossRef]
- Su, S.; Wong, G.; Shi, W.; Liu, J.; Lai, A.C.K.; Zhou, J.; Liu, W.; Bi, Y.; Gao, G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016, 24, 490–502. [Google Scholar] [CrossRef]
- Quintana, H.; Pacheco, V. Identificación y Distribución de los Murciélagos Vampiros del Perú; Lima. Rev. Perú. Med. Exp. Salud Publica 2007, 24, 81–88. Available online: http://www.scielo.org.pe/scielo.php?script=sci_arttext&pid=S1726-46342007000100011 (accessed on 1 August 2023).
- Bergner, L.M.; Orton, R.J.; da Silva Filipe, A.; Shaw, A.E.; Becker, D.J.; Tello, C.; Biek, R.; Streicker, D.G. Using noninvasive metagenomics to characterize viral communities from wildlife. Mol. Ecol. Resour. 2019, 19, 128–143. [Google Scholar] [CrossRef]
- Simas, P.V.M.; Barnabé, A.C.S.; Durães-Carvalho, R.; Neto, D.F.L.; Caserta, L.C.; Artacho, L.; Jacomassa, F.A.F.; Martini, M.C.; Bianchi dos Santos, M.M.A.; Felippe, P.A.N.; et al. Bat coronavirus in Brazil related to appalachian ridge and porcine epidemic diarrhea viruses. Emerg. Infect. Dis. 2015, 21, 729–731. [Google Scholar] [CrossRef]
- Ahmad, T.; Khan, M.; Haroon; Musa, T.H.; Nasir, S.; Hui, J.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. COVID-19: Zoonotic Aspects. Travel Med. Infect. Dis. 2020, 36, 101607. [Google Scholar] [CrossRef]
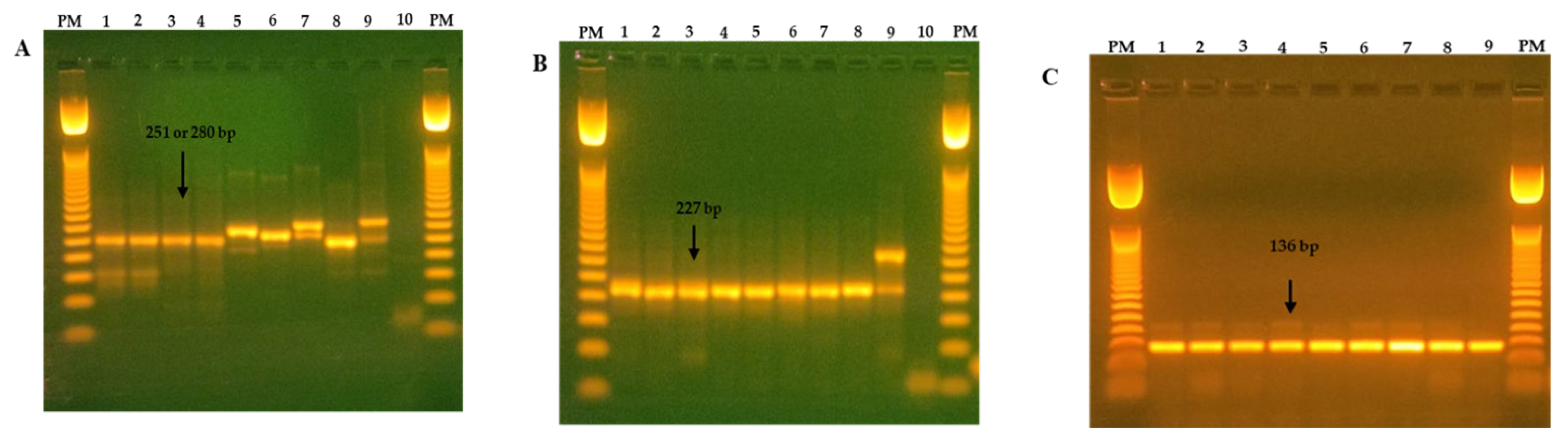


| Virus | Gene | Assay | Primer * | Primer Sequence 5′ → 3′ | Position | Product Size (bp) | Reference |
|---|---|---|---|---|---|---|---|
| All CoV | RdRd + | RT-PCR | Cor-FW | ACWCARHTVAAYYTNAARTAYGC | 14,922–14,944 | 251 | [41] |
| Cor-RV | TCRCAYTTDGGRTARTCCCA | 15,153–15,172 | |||||
| β-CoV | RdRd | Nested PCR | Beta.CoV.F | ATTAGTGCWAAGAATAGAGCYCGCAC | 14,946–14,971 | 227 | [9] |
| Beta.CoV.R | TCACAYTTWGGRTARTCCCADCCCA | 15,148–15,172 | |||||
| Embecovirus | RdRd | Nested PCR | CV2U.F | TACTATGACTGGCAGAATGTTTCA | 14,996–15,019 | 136 | [42] |
| CV2L.R | AACATCTTTAATAAGGCGRCGTAA | 15,108–15,131 |
| Infection Type | Genus | Subgenus | Nº Positive Samples |
|---|---|---|---|
| Single detection | Betacoronavirus (β-CoV) | Embecovirus (EmbeCoV) | 15 |
| Not identified | 9 | ||
| Alphacoronavirus (α-CoV) | Decacovirus (DecaCoV) | 1 | |
| Unclassified (Megaderma Bat-CoV-like) | Unclassified (Megaderma Bat-CoV-like) | 1 | |
| Not identified | Not identified | 2 | |
| Multiple detection | β-CoV + α-CoV | EmbeCoV + DecaCoV | 1 |
| β-CoV + Unclassified (Megaderma Bat-CoV-like) | EmbeCoV + unclassified | 1 | |
| Total | 30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llanco, L.; Retamozo, K.; Oviedo, N.; Manchego, A.; Lázaro, C.; Navarro-Mamani, D.A.; Santos, N.; Rojas, M. Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas. Animals 2023, 13, 2983. https://doi.org/10.3390/ani13182983
Llanco L, Retamozo K, Oviedo N, Manchego A, Lázaro C, Navarro-Mamani DA, Santos N, Rojas M. Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas. Animals. 2023; 13(18):2983. https://doi.org/10.3390/ani13182983
Chicago/Turabian StyleLlanco, Luis, Karubya Retamozo, Noriko Oviedo, Alberto Manchego, César Lázaro, Dennis A. Navarro-Mamani, Norma Santos, and Miguel Rojas. 2023. "Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas" Animals 13, no. 18: 2983. https://doi.org/10.3390/ani13182983
APA StyleLlanco, L., Retamozo, K., Oviedo, N., Manchego, A., Lázaro, C., Navarro-Mamani, D. A., Santos, N., & Rojas, M. (2023). Co-Circulation of Multiple Coronavirus Genera and Subgenera during an Epizootic of Lethal Respiratory Disease in Newborn Alpacas (Vicugna pacos) in Peru: First Report of Bat-like Coronaviruses in Alpacas. Animals, 13(18), 2983. https://doi.org/10.3390/ani13182983







