The Distribution of IgT mRNA+ Cells in the Gut of the Atlantic Salmon (Salmo salar L.)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Study
2.2. Sampling
2.3. In Situ Hybridization
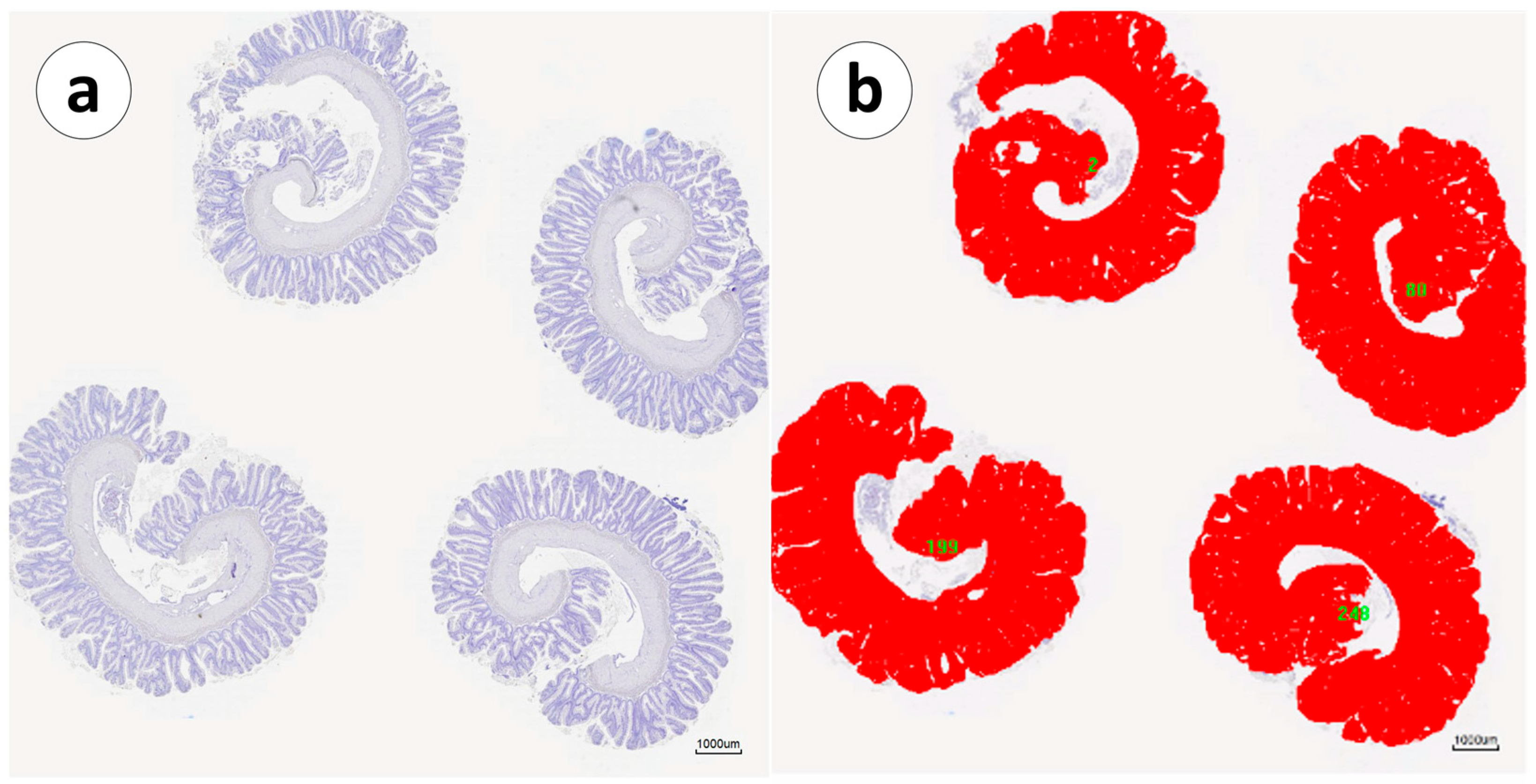
2.4. Image Acquisition and mRNA+ Cell Counting
2.5. Statistical Analyses
2.6. Ethical Statement
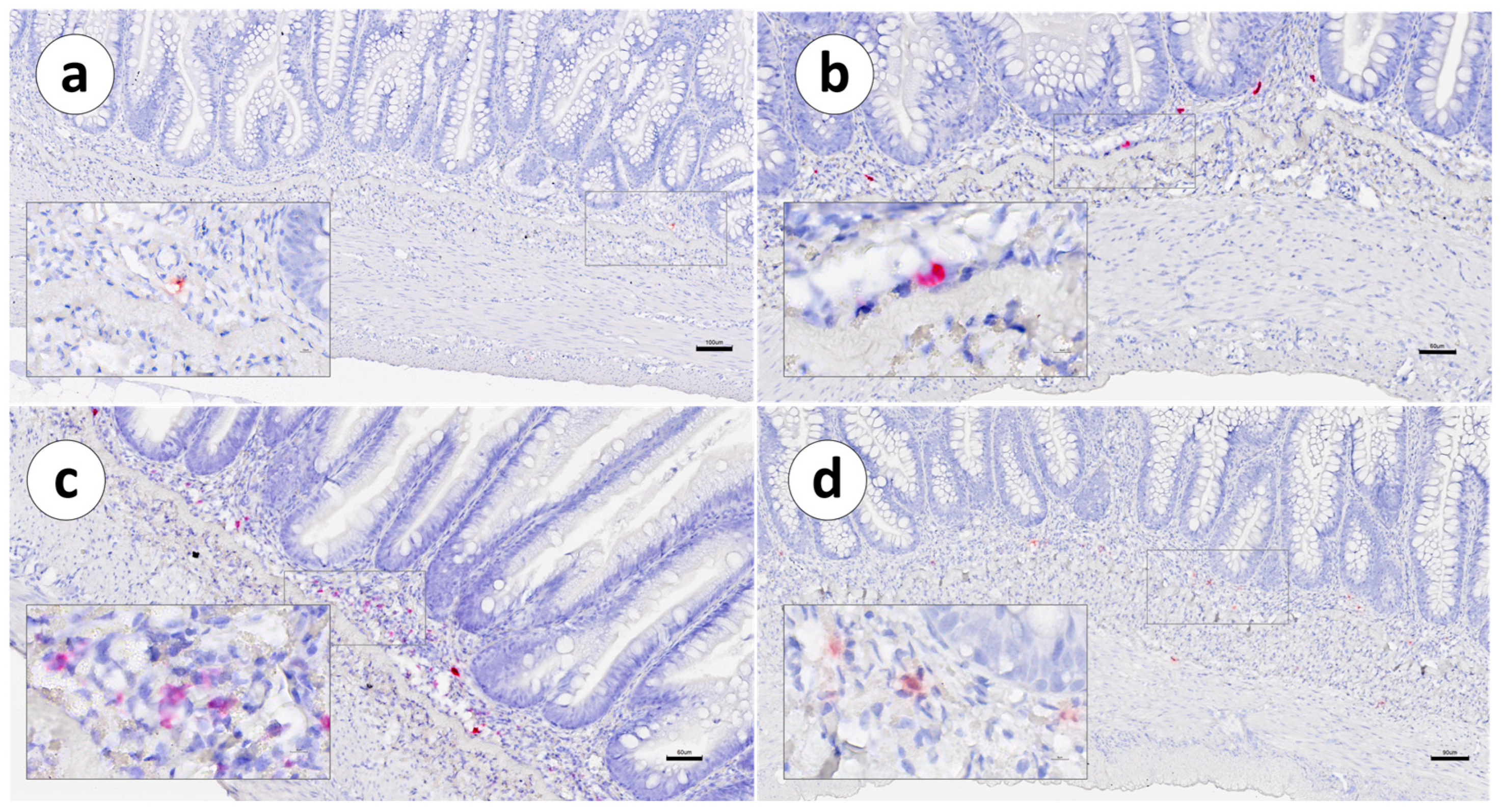
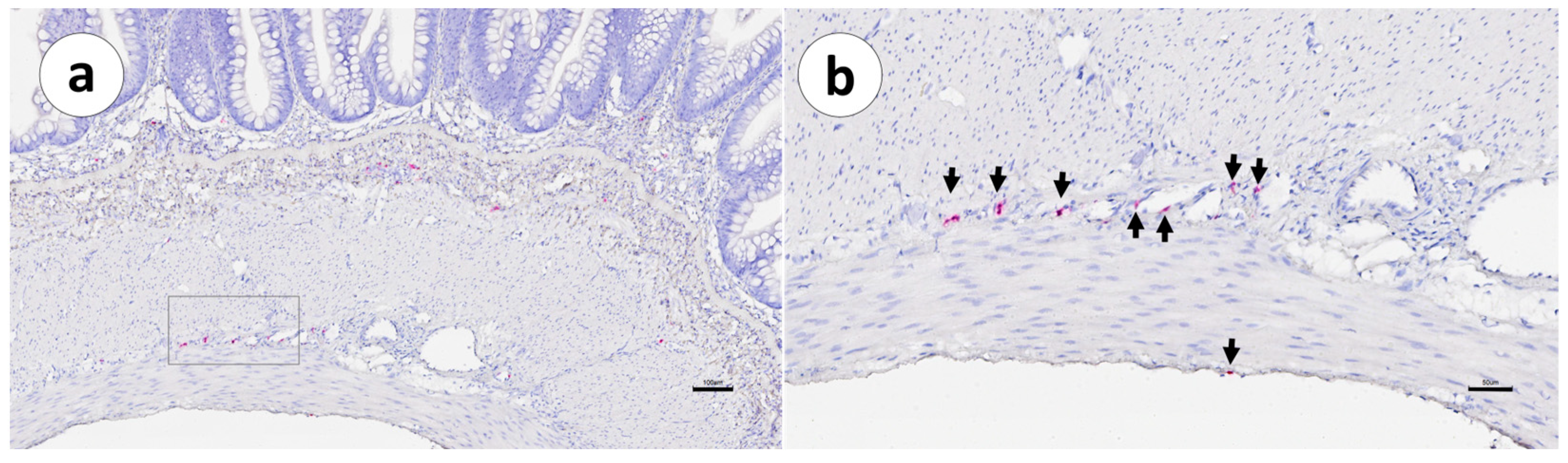
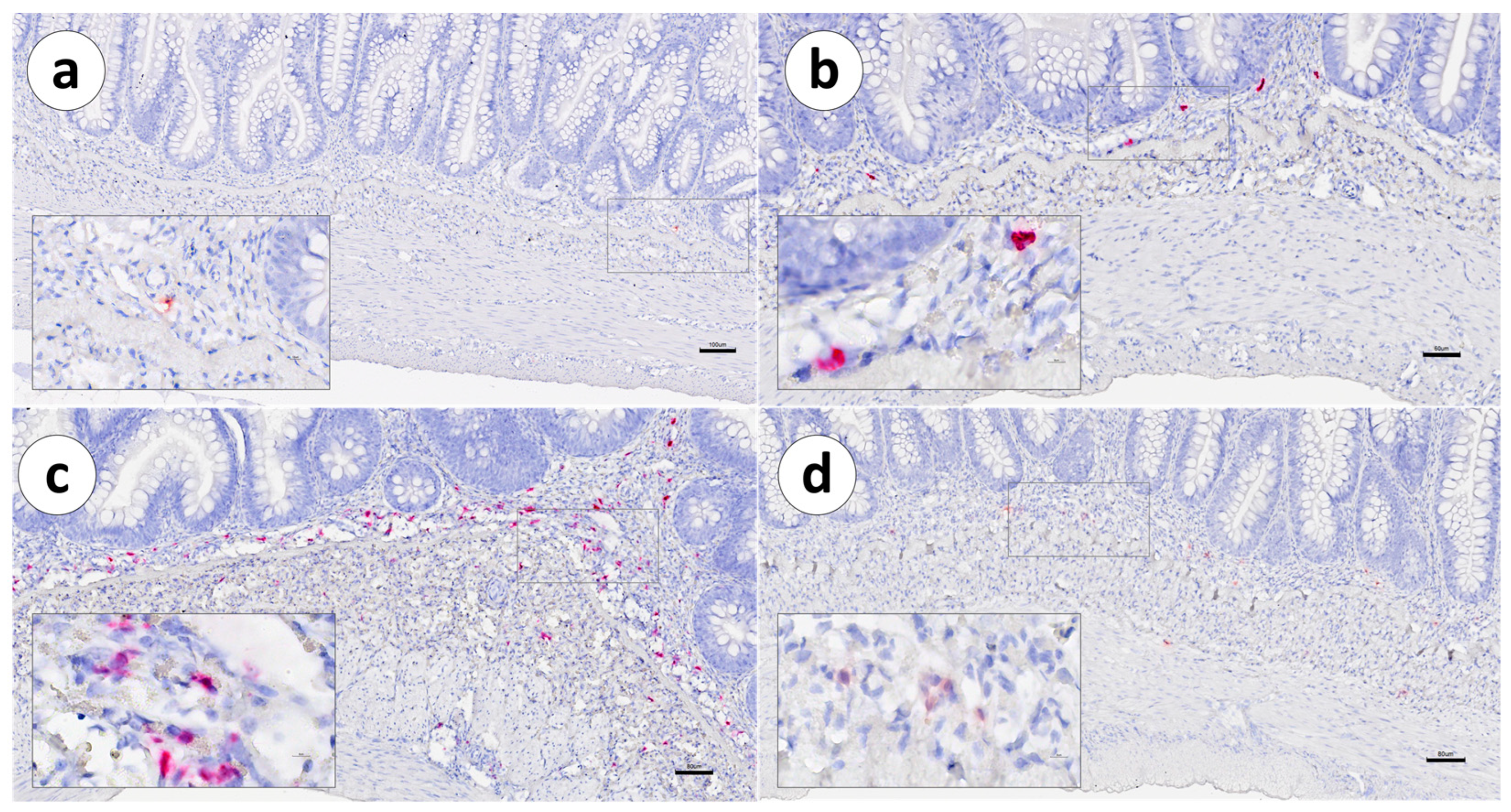
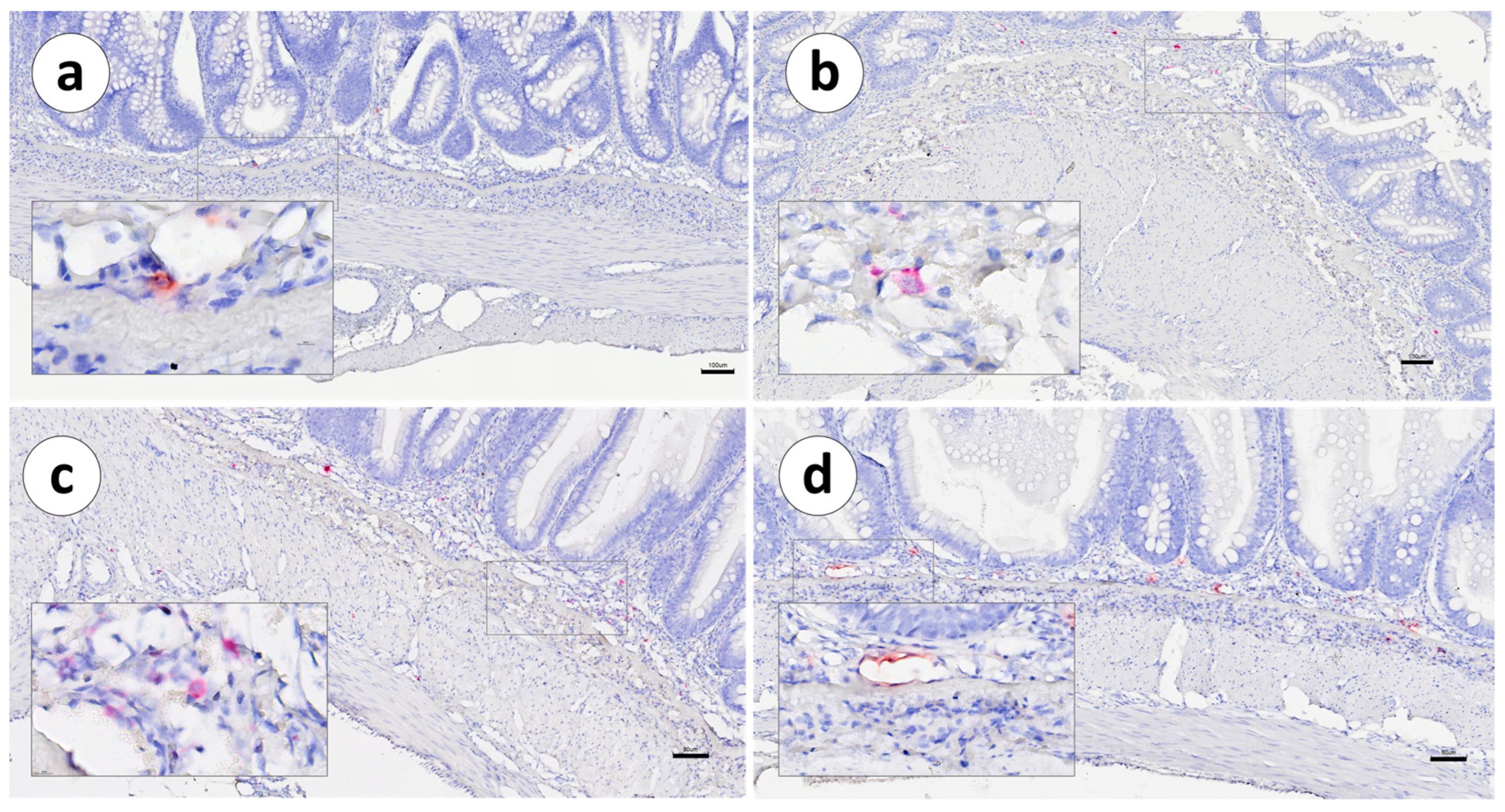
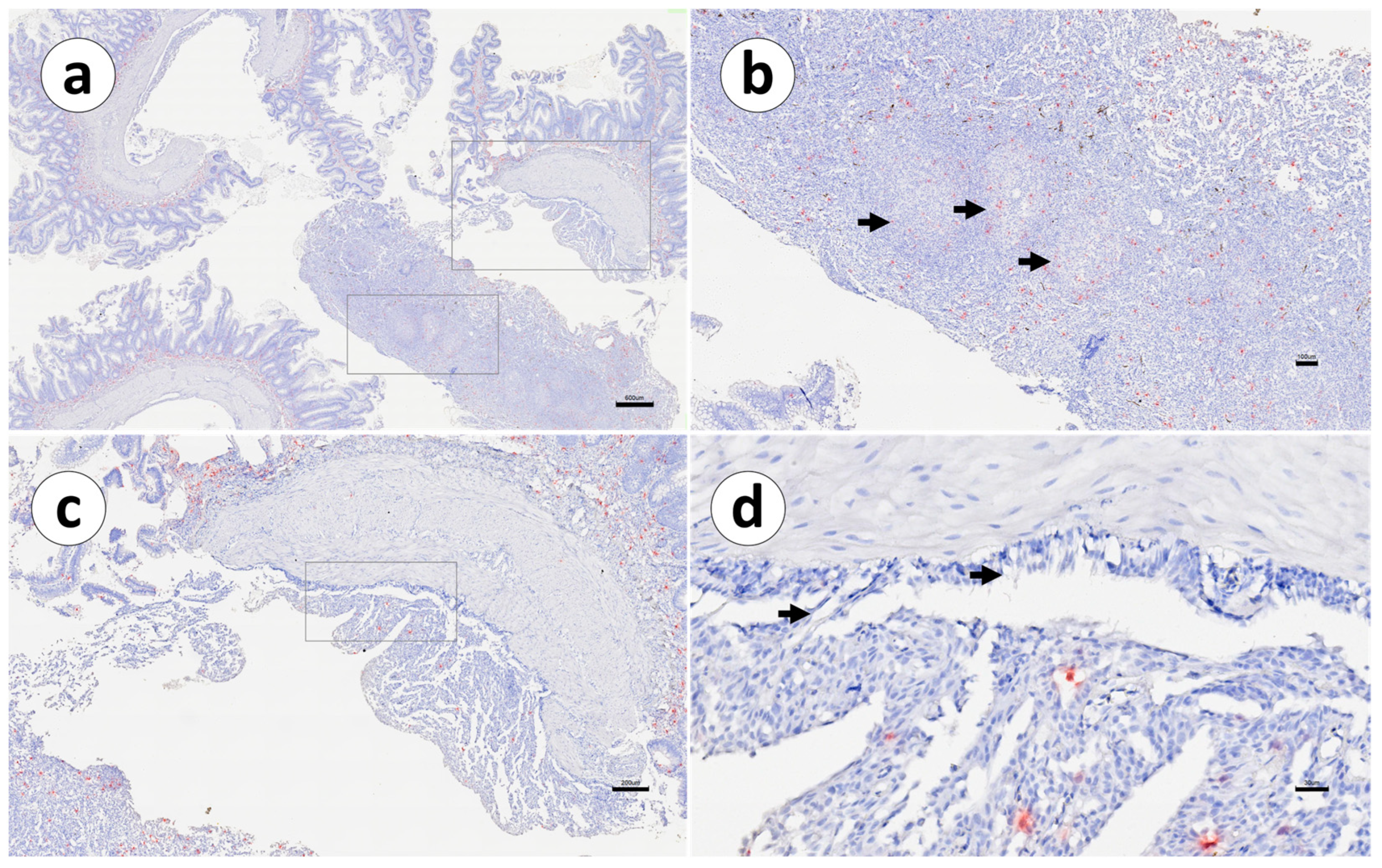
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bilal, S.; Etayo, A.; Hordvik, I. Immunoglobulins in teleosts. Immunogenetics 2021, 73, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.A.; Salinas, I.; Li, J.; Parra, D.; Bjork, S.; Xu, Z.; Lapatra, S.E.; Bartholomew, J.; Sunyer, J.O. IgT, a primitive immu-noglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef]
- Zhang, Y.A.; Salinas, I.; Oriol Sunyer, J. Recent findings on the structure and function of teleost IgT. Fish Shellfish Immunol. 2011, 31, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.Y.; Kong, W.; Yin, Y.X.; Dong, F.; Huang, Z.Y.; Yin, G.M.; Dong, S.; Salinas, I.; Zhang, Y.A.; Xu, Z. Mucosal immuno-globulins protect the olfactory organ of teleost fish against parasitic infection. PLoS Pathog. 2018, 14, e1007251. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Jouneau, L.; Pham, H.-P.; Bouchez, O.; Ronique Giudicelli, V.; Lefranc, M.-P.; Quillet, E.; Benmansour, A.; Dé, F.; Cazals, R.; et al. Teleost Fish Mount Complex Clonal IgM and IgT Responses in Spleen upon Systemic Viral Infection. PLoS Pathog. 2013, 9, e1003098. [Google Scholar] [CrossRef]
- Buonocore, F.; Stocchi, V.; Nunez-Ortiz, N.; Randelli, E.; Gerdol, M.; Pallavicini, A.; Facchiano, A.; Bernini, C.; Guerra, L.; Scapigliati, G.; et al. Immunoglobulin T from sea bass (Dicentrarchus labrax L.): Molecular characterization, tissue localization and expression after nodavirus infection. BMC Mol. Biol. 2017, 18, 8. [Google Scholar] [CrossRef]
- Makesh, M.; Sudheesh, P.S.; Cain, K.D. Systemic and mucosal immune response of rainbow trout to immunization with an attenuated flavobacterium psychrophilum vaccine strain by different routes. Fish Shellfish Immunol. 2015, 44, 156–163. [Google Scholar] [CrossRef]
- Gao, C.; Fu, Q.; Su, B.; Zhou, S.; Liu, F.; Song, L.; Zhang, M.; Ren, Y.; Dong, X.; Tan, F.; et al. Transcriptomic profiling revealed the signatures of intestinal barrier alteration and pathogen entry in turbot (Scophthalmus maximus) following Vibrio anguillarum challenge. Dev. Comp. Immunol. 2016, 65, 159–168. [Google Scholar] [CrossRef]
- Salinas, I.; Parra, D. Fish mucosal immunity: Intestine. In Mucosal Health in Aquaculture; Elsevier: Amsterdam, The Netherlands, 2015; pp. 135–170. [Google Scholar] [CrossRef]
- Bjørgen, H.; Hellberg, H.; Løken, O.M.; Gunnes, G.; Koppang, E.O.; Dale, O.B. Tumor microenvironment and stroma in in-testinal adenocarcinomas and associated metastases in Atlantic salmon broodfish (Salmo salar). Vet. Immunol. Immunopathol. 2019, 214, 109891. [Google Scholar] [CrossRef]
- Ballesteros, N.A.; Castro, R.; Abos, B.; Saint-Jean, R.; Pérez-Prieto, S.S. The Pyloric Caeca Area Is a Major Site for IgM + and IgT + B Cell Recruitment in Response to Oral Vaccination in Rainbow Trout. PLoS ONE 2013, 8, 66118. [Google Scholar] [CrossRef] [PubMed]
- Løkka, G.; Austbø, L.; Falk, K.; Bromage, E.; Fjelldal, P.G.; Hansen, T.; Hordvik, I.; Koppang, E.O. Immune parameters in the intestine of wild and reared unvaccinated and vaccinated Atlantic salmon (Salmo salar L.). Dev. Comp. Immunol. 2014, 47, 6–16. [Google Scholar] [CrossRef]
- Bjørgen, H.; Koppang, E.O. Anatomy of Teleost Fish Immune Structures and Organs. In Principles of Fish Immunology; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–30. [Google Scholar] [CrossRef]
- Bjoergen, H.; Hordvik, I.; Koppang, E.O. Inflammatory changes and melanization in the Atlantic salmon (Salmo salar) targeted by in situ hybridization. J. Immunol. 2019, 202, 73.6. [Google Scholar] [CrossRef]
- Jirapongpairoj, W.; Hirono, I.; Kondo, H. Development and evaluation of polyclonal antisera for detection of the IgM heavy chain of multiple fish species. J. Immunol. Methods 2017, 449, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Li, L.; Duan, C.; Wu, L.; Tu, X.; Vogelbein, M.A.; Bromage, E.; Kaattari, S.L. IgM-bearing B cell affinity subpopulations possess differential antigen sensitivity in rainbow trout. Fish Shellfish Immunol. 2021, 118, 111–118. [Google Scholar] [CrossRef]
- Danilova, N.; Bussmann, J.; Jekosch, K.; Steiner, L.A. The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z. Nat. Immunol. 2005, 6, 295–302. [Google Scholar] [CrossRef]
- Hansen, J.D.; Landis, E.D.; Phillips, R.B. Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish. Proc. Natl. Acad. Sci. USA 2005, 102, 6919–6924. [Google Scholar] [CrossRef]
- Mobley, K.B.; Aykanat, T.; Czorlich, Y.; House, A.; Kurko, J.; Miettinen, A.; Moustakas-Verho, J.; Salgado, A.; Sinclair-Waters, M.; Verta, J.P.; et al. Maturation in Atlantic salmon (Salmo salar, Salmonidae): A synthesis of ecological, genetic, and molecular processes. Rev. Fish Biol. Fish. 2021, 31, 523–571. [Google Scholar] [CrossRef]
- Yasuike, M.; de Boer, J.; von Schalburg, K.R.; Cooper, G.A.; McKinnel, L.; Messmer, A.; So, S.; Davidson, W.S.; Koop, B.F. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genom. 2010, 11, 486. [Google Scholar] [CrossRef]
- Xia, H.; Yang, P.; Chen, Z.; Zhang, Y.; Liu, L.; Luo, Y.; Meng, S.; Fang, X.; Yuan, M.; Liu, Y.; et al. Polymeric immunoglobulin receptor in dongtingking crucian carp (Carassius auratus indigentiaus): Molecular characterization and expression analysis in response to Aeromonas hydrophila challenge. Aquac. Res. 2022, 53, 3818–3827. [Google Scholar] [CrossRef]
- Etayo, A.; Bjørgen, H.; Koppang, E.O.; Hordvik, I. The teleost polymeric Ig receptor counterpart in ballan wrasse (Labrus bergylta) differs from pIgR in higher vertebrates. Vet. Immunol. Immunopathol. 2022, 249, 110440. [Google Scholar] [CrossRef]
- Yang, D.; Hu, X.; Li, H.; Xu, W.; Wu, T.; Chen, J. Molecular cloning and characteristic analysis of polymeric immunoglobulin receptor-like (plgRL) in large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol. 2023, 132, 108503. [Google Scholar] [CrossRef] [PubMed]
- Løkka, G.; Austb, L.; Falk, K.; Bjerkås, I.; Koppang, E.O. Intestinal morphology of the wild atlantic salmon (Salmo salar). J. Morphol. 2013, 274, 859–876. [Google Scholar] [CrossRef] [PubMed]
- Løken, O.M.; Bjørgen, H.; Hordvik, I.; Koppang, E.O. A teleost structural analogue to the avian bursa of Fabricius. J. Anat. 2020, 236, 798–808. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Flanagan, J.; Su, N.; Wang, L.-C.; Bui, S.; Nielson, A.; Wu, X.; Vo, H.-T.; Ma, X.-J.; Luo, Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 2012, 14, 22–29. [Google Scholar] [CrossRef]
- Koppang, E.O.; Lundin, M.; Press, C.M.; Rønningen, K.; Lie, Ø. Differing levels of Mhc class II β chain expression in a range of tissues from vaccinated and non-vaccinated Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 1998, 8, 183–196. [Google Scholar] [CrossRef]
- Bjørgen, H.; Koppang, E.O.; Gunnes, G.; Hordvik, I.; Moldal, T.; Kaldhusdal, M.; Dale, O.B. Ectopic epithelial cell clusters in salmonid intestine are associated with inflammation. J. Fish Dis. 2018, 41, 1031–1040. [Google Scholar] [CrossRef]
- Bilal, S.; Lie, K.K.; Dalum, A.S.; Karlsen, O.A.; Hordvik, I. Analysis of immunoglobulin and T cell receptor gene expression in ballan wrasse (Labrus bergylta) revealed an extraordinarily high IgM expression in the gut. Fish Shellfish Immunol. 2019, 87, 650–658. [Google Scholar] [CrossRef]
- Fuglem, B.; Jirillo, E.; Bjerkås, I.; Kiyono, H.; Nochi, T.; Yuki, Y.; Raida, M.; Fischer, U.; Koppang, E.O. Antigen-sampling cells in the salmonid intestinal epithelium. Dev. Comp. Immunol. 2010, 34, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.N.; Akhter, S.; Smith, E.M.; Lorent, K.; Pack, M. Intestinal growth and differentiation in zebrafish. Mech. Dev. 2005, 122, 157–173. [Google Scholar] [CrossRef]
- Hrubec, T.C.; Ward, D.; Smith, S.A.; Robertson, J.L. Age related changes in humoral immune response of hybrid striped bass (Morone chrysops × Morone saxatilis). Vet. Immunol. Immunopathol. 2004, 101, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Fuda, H.; Soyano, K.; Yamazaki, F.; Hara, A. Serum immunoglobulin M (IgM) during early development of masu salmon (Oncorhynchus masou). Comp. Biochem. Physiol.-A Comp. Physiol. 1991, 99, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Nagae, M.; Fuda, H.; Hara, A.; Kawamura, H.; Yamauchi, K. Changes in serum immunoglobulin M (IgM) con-centrations during early development of chum salmon (Oncorhynchus keta) as determined by sensitive ELISA technique. Comp. Biochem. Physiol. Part A Physiol. 1993, 106, 69–74. [Google Scholar] [CrossRef]
- Melingen, G.O.; Stefansson, S.O.; Berg, A.; Wergeland, H.I. Changes in serum protein and IgM concentration during smolting and early post-smolt period in vaccinated and unvaccinated Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 1995, 5, 211–221. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, X.; Yu, Z.; Sun, Z.; Li, J.; Guan, C.; Lei, J.; Ma, A.; Shan, H. Expression and localization study of pIgR in the late stage of embryo development in turbot (Scophthalmus maximus). Fish Shellfish Immunol. 2019, 87, 315–321. [Google Scholar] [CrossRef]
- Kong, X.; Wang, L.; Pei, C.; Zhang, J.; Zhao, X.; Li, L. Comparison of polymeric immunoglobulin receptor between fish and mammals. Vet. Immunol. Immunopathol. 2018, 202, 63–69. [Google Scholar] [CrossRef]
- Peppard, J.V.; Jackson, L.E.; Hall, J.G.; Robertson, D. The transfer of immune complexes from the lumen of the small intestine to the bloodstream in sucking rats. Immunology 1984, 53, 385–393. [Google Scholar]
- Johansen, F.E.; Kaetzel, C.S. Regulation of the polymeric immunoglobulin receptor and IgA transport: New advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 2011, 4, 598–602. [Google Scholar] [CrossRef]
- Xu, Z.; Takizawa, F.; Casadei, E.; Shibasaki, Y.; Ding, Y.; Sauters, T.J.C.; Yu, Y.; Salinas, I.; Sunyer, J.O. Specialization of mu-cosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol. 2020, 5, eaay3254. [Google Scholar] [CrossRef]
- Midtlyng, P.J.; Reitan, L.J.; Speilberg, L. Experimental studies on the efficacy and side-effects of intraperitoneal vaccination of Atlantic salmon (Salmo salar L.) against furunculosis. Fish Shellfish Immunol. 1996, 6, 335–350. [Google Scholar] [CrossRef]
- Haramati, N.; Lorans, R.; Lutwin, M.; Kaleya, R.N. Injection granulomas. Intramuscle or intrafat? Arch. Fam. Med. 1994, 3, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Shintani, Y.; Fields, L.; Shiraishi, M.; Podaru, M.N.; Kainuma, S.; Yamashita, K.; Kobayashi, K.; Perretti, M.; Lewis-McDougall, F.; et al. Cell barrier function of resident peritoneal macrophages in post-operative adhesions. Nat. Commun. 2021, 12, 2232. [Google Scholar] [CrossRef] [PubMed]
- Okabe, Y.; Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 2014, 157, 832–844. [Google Scholar] [CrossRef] [PubMed]
- Maruya, M.; Suzuki, K.; Fujimoto, H.; Miyajima, M.; Kanagawa, O.; Wakayama, T.; Fagarasan, S. Vitamin A-dependent transcriptional activation of the nuclear factor of activated T cells c1 (NFATc1) is critical for the development and survival of B1 cells. Proc. Natl. Acad. Sci. USA 2011, 108, 722–727. [Google Scholar] [CrossRef] [PubMed]
- Ha, S.A.; Tsuji, M.; Suzuki, K.; Meek, B.; Yasuda, N.; Kaisho, T.; Fagarasan, S. Regulation of B1 cell migration by signals through Toll-like receptors. J. Exp. Med. 2006, 203, 2541–2550. [Google Scholar] [CrossRef] [PubMed]
- Jenberie, S. Atlantic Salmon B Cells-Local and Systemic Responses to Intraperitoneally Administered Salmonid Alphavirus. Ph.D. Thesis, The Arctic University of Norway, Tromsø, Norway, 4 March 2020. Available online: https://hdl.handle.net/10037/17566 (accessed on 4 May 2023).








| Group 1 (avg. 153 g) | Group 2 (avg. 1717 g) | ||
|---|---|---|---|
| Unvaccinated | Vaccinated | Unvaccinated | Vaccinated |
| (3 fish) | (3 fish) | (3 fish) | (3 fish) |
| Probe | Accession No. | Target Region (bp) | Catalog No. | |
|---|---|---|---|---|
| Target | IgT | GQ907003 | 3–883 | 532171 |
| pIgR | GQ892057.1 | 119–1145 | 845451 | |
| Control | dapB (negative) | EF191515 | 414–862 | 310043 |
| ppib (positive) | NM_001140870 | 20–934 | 494421 |
| Probe | Section | Group 1 | Group 2 | ||
|---|---|---|---|---|---|
| Unvaccinated | Vaccinated | Unvaccinated | Vaccinated | ||
| IgT | Pc | − | − | − | + |
| FsMi | − | − | − | + | |
| SsMi | −/+ | −/+ | −/+ | ++ | |
| Ps | − | − | − | + | |
| pIgR | SsMi | + | −/+ | + | −/+ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, P.L.; Barac, F.; Hansen, T.J.; Fjelldal, P.G.; Hordvik, I.; Bjørgen, H.; Koppang, E.O. The Distribution of IgT mRNA+ Cells in the Gut of the Atlantic Salmon (Salmo salar L.). Animals 2023, 13, 3191. https://doi.org/10.3390/ani13203191
Castro PL, Barac F, Hansen TJ, Fjelldal PG, Hordvik I, Bjørgen H, Koppang EO. The Distribution of IgT mRNA+ Cells in the Gut of the Atlantic Salmon (Salmo salar L.). Animals. 2023; 13(20):3191. https://doi.org/10.3390/ani13203191
Chicago/Turabian StyleCastro, Pedro Luis, Fran Barac, Tom Johnny Hansen, Per Gunnar Fjelldal, Ivar Hordvik, Håvard Bjørgen, and Erling Olaf Koppang. 2023. "The Distribution of IgT mRNA+ Cells in the Gut of the Atlantic Salmon (Salmo salar L.)" Animals 13, no. 20: 3191. https://doi.org/10.3390/ani13203191
APA StyleCastro, P. L., Barac, F., Hansen, T. J., Fjelldal, P. G., Hordvik, I., Bjørgen, H., & Koppang, E. O. (2023). The Distribution of IgT mRNA+ Cells in the Gut of the Atlantic Salmon (Salmo salar L.). Animals, 13(20), 3191. https://doi.org/10.3390/ani13203191






