Assessing the Seasonal and Spatial Dynamics of Zooplankton through DNA Metabarcoding in a Temperate Estuary
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
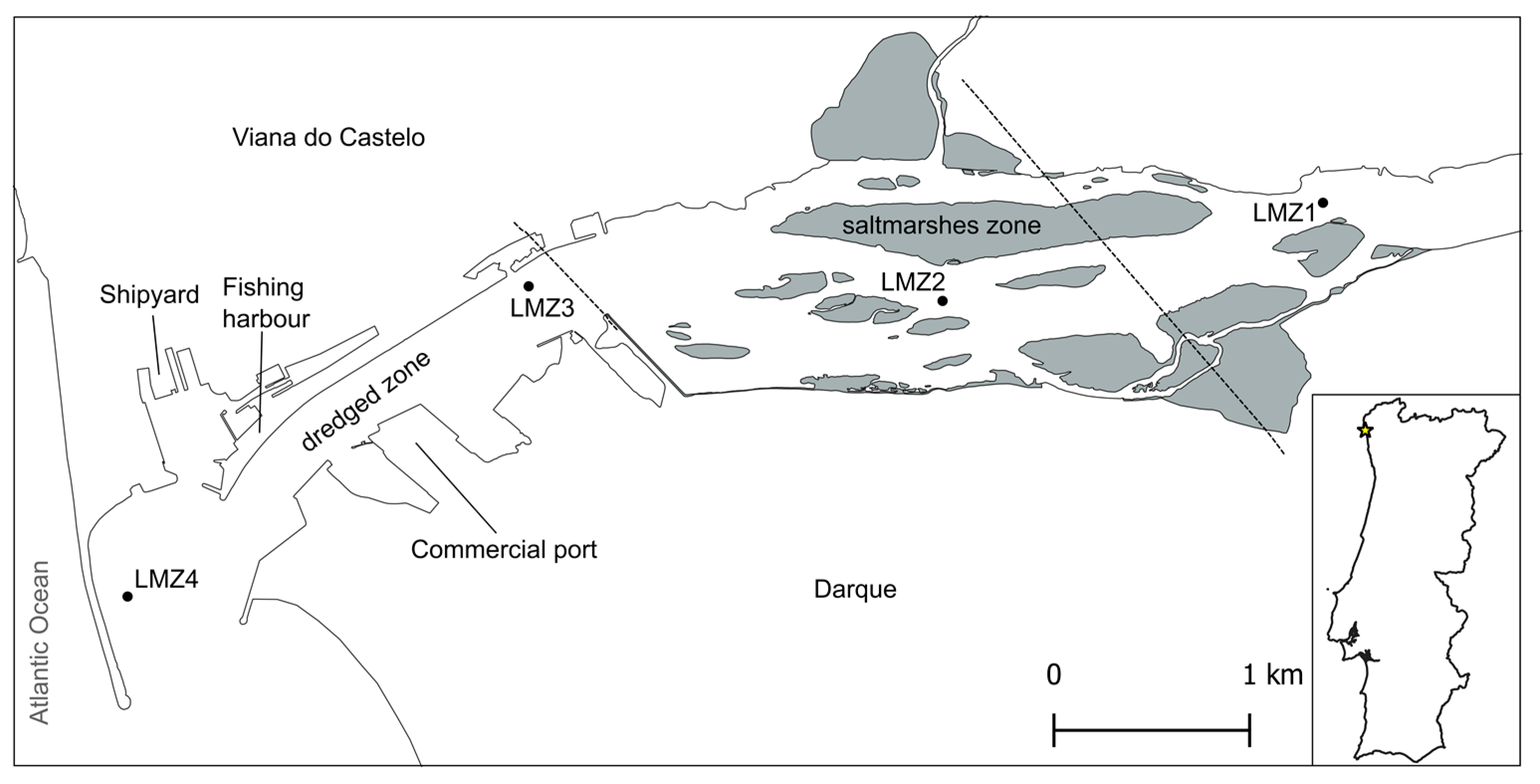
2.1. Description of the Study Area
2.2. Sampling Strategy
2.3. DNA Extraction, PCR Amplification, and High-Throughput Sequencing (HTS)
2.4. Bioinformatic Processing and Taxonomic Assignment
2.5. Data Processing and Analysis
3. Results
3.1. HTS Data Initial Processing and the General Taxonomic Composition Recovered by Each Marker
3.2. Non-Indigenous Species Detection
3.3. Seasonal and Spatial Dynamic Effects on Species Richness, Taxonomic Composition, and Distinctness
3.4. Seasonal and Spatial Dynamics of Zooplankton Structure Composition
4. Discussion
4.1. DNA Metabarcoding Performance in the Assessment of Zooplankton Species in the Lima Estuary
4.2. COI and 18S rRNA Gene Markers Displayed Minute Overlap in Zooplankton Species Detection
4.3. DNA Metabarcoding Performance in the Detection of Non-Indigenous Species (NIS) in Zooplankton Samples
4.4. Species Richness, Taxonomic Distinctness, and Species Composition Influenced Primarly by Season and Secondarily by Within-Estuary Location
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hellawell, J.M. Biological Indicators of Freshwaters Pollution and Environmental Management; Mellanby, K., Ed.; Elsevier: London, UK; New York, NY, USA, 1986; ISBN 9789400943155. [Google Scholar]
- Morse, J.C.; Bae, Y.J.; Munkhjargal, G.; Sangpradub, N.; Tanida, K.; Vshivkova, T.S.; Wang, B.; Yang, L.; Yule, C.M. Freshwater Biomonitoring with Macroinvertebrates in East Asia. Front. Ecol. Environ. 2007, 5, 33–42. [Google Scholar] [CrossRef]
- Ruaro, R.; Gubiani, É.A.; Cunico, A.M.; Moretto, Y.; Piana, P.A. Comparison of Fish and Macroinvertebrates as Bioindicators of Neotropical Streams. Environ. Monit. Assess. 2016, 188, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Caroni, R.; Irvine, K. The Potential of Zooplankton Communities for Ecological Assessment of Lakes: Redundant Concept or Political Oversight? Biol. Environ. 2010, 110, 35–53. [Google Scholar] [CrossRef]
- Landry, M.R.; Hassett, R.P. Estimating the Grazing Impact of Marine Micro-Zooplankton. Mar. Biol. 1982, 67, 283–288. [Google Scholar] [CrossRef]
- Ikeda, T. Nutritional Ecology of Zooplankton. Ph.D. Thesis, Hokkaido University, Hakodate, Japan, 1974. [Google Scholar]
- Suthers, I.M.; Rissik, D. Plankton: A Guide to Their Ecology and Monitoring for Water Quality, 2nd ed.; CSIRO: Canberra, Australia, 2009; ISBN 9780643090583. [Google Scholar]
- Le Quéré, C.; Buitenhuis, E.T.; Moriarty, R.; Alvain, S.; Aumont, O.; Bopp, L.; Chollet, S.; Enright, C.; Franklin, D.J.; Geider, R.J.; et al. Role of Zooplankton Dynamics for Southern Ocean Phytoplankton Biomass and Global Biogeochemical Cycles. Biogeosciences 2016, 13, 4111–4133. [Google Scholar] [CrossRef]
- Steinberg, D. Zooplankton Biogeochemical Cycles. In Marine Plankton: A Practical Guide to Ecology, Methodology, and Taxonomy; Castellani, C., Edwards Martin, Eds.; Oxford University Press: Oxford, UK, 2017; Volume 1. [Google Scholar]
- Gannon, J.E.; Stemberger, R.S. Zooplankton (Especially Crustaceans and Rotifers) as Indicators of Water Quality. Trans. Am. Microsc. Soc. 1978, 97, 16–35. [Google Scholar] [CrossRef]
- An, X.P.; Du, Z.H.; Zhang, J.H.; Li, Y.P.; Qi, J.W. Structure of the Zooplankton Community in Hulun Lake, China. Procedia Environ. Sci. 2012, 13, 1099–1109. [Google Scholar] [CrossRef]
- Azevêdo, D.J.S.; Barbosa, J.E.L.; Gomes, W.I.A.; Porto, D.E.; Marques, J.C.; Molozzi, J. Diversity Measures in Macroinvertebrate and Zooplankton Communities Related to the Trophic Status of Subtropical Reservoirs: Contradictory or Complementary Responses? Ecol. Indic. 2015, 50, 135–149. [Google Scholar] [CrossRef]
- Kour, S.; Slathia, D.; Sharma, N.; Kour, S.; Verma, R. Zooplankton as Bioindicators of Trophic Status of a Lentic Water Source, Jammu (J&K) with Remarks on First Reports. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2022, 92, 393–404. [Google Scholar] [CrossRef]
- Chiba, S.; Batten, S.; Martin, C.S.; Ivory, S.; Miloslavich, P.; Weatherdon, L.V. Zooplankton Monitoring to Contribute towards Addressing Global Biodiversity Conservation Challenges. J. Plankton Res. 2018, 40, 509–518. [Google Scholar] [CrossRef]
- Almeida, R.; Formigo, N.E.; Sousa-Pinto, I.; Antunes, S.C. Contribution of Zooplankton as a Biological Element in the Assessment of Reservoir Water Quality. Limnetica 2020, 39, 245–261. [Google Scholar] [CrossRef]
- Vieira, L.R.; Guilhermino, L.; Morgado, F. Zooplankton Structure and Dynamics in Two Estuaries from the Atlantic Coast in Relation to Multi-Stressors Exposure. Estuar. Coast. Shelf Sci. 2015, 167, 347–367. [Google Scholar] [CrossRef]
- Berasategui, A.A.; Calliari, D.L.; Amodeo, M.; Spetter, C.V.; Guinder, V.; Biancalana, F. Interannual Changes in Winter-Spring Zooplankton Estuarine Community Forced by Hydroclimatic Variability—With Special Reference to Bioindicator Species Eurytemora Americana. Mar. Environ. Res. 2023, 186, 105898. [Google Scholar] [CrossRef]
- Marques, S.C.; Pardal, M.Â.; Primo, A.L.; Martinho, F.; Falcão, J.; Azeiteiro, U.; Molinero, J.C. Evidence for Changes in Estuarine Zooplankton Fostered by Increased Climate Variance. Ecosystems 2018, 21, 56–67. [Google Scholar] [CrossRef]
- Almeida, L.R.; Costa, I.S.; Eskinazi-Sant’Anna, E.M. Composition and Abundance of Zooplankton Community of an Impacted Estuarine Lagoon in Northeast Brazil. Braz. J. Biol. 2012, 72, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Goswami, S.C. Zooplankton Methodology, Collection & Identification—A Field Manual, 1st ed.; Dhargalkar, V.K., Verlecar, X.N., Eds.; National Institute of Oceanography: Goa, India, 2004. [Google Scholar]
- Jeppesen, E.; Nõges, P.; Davidson, T.A.; Haberman, J.; Nõges, T.; Blank, K.; Lauridsen, T.L.; Søndergaard, M.; Sayer, C.; Laugaste, R.; et al. Zooplankton as Indicators in Lakes: A Scientific-Based Plea for Including Zooplankton in the Ecological Quality Assessment of Lakes According to the European Water Framework Directive (WFD). Hydrobiologia 2011, 676, 279–297. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards Next-Generation Biodiversity Assessment Using DNA Metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef]
- Creer, S.; Deiner, K.; Frey, S.; Porazinska, D.; Taberlet, P.; Thomas, W.K.; Potter, C.; Bik, H.M. The Ecologist’s Field Guide to Sequence-Based Identification of Biodiversity. Methods Ecol. Evol. 2016, 7, 1008–1018. [Google Scholar] [CrossRef]
- Shokralla, S.; Spall, J.L.; Gibson, J.F.; Hajibabaei, M. Next-Generation Sequencing Technologies for Environmental DNA Research. Mol. Ecol. 2012, 21, 1794–1805. [Google Scholar] [CrossRef]
- Bucklin, A.; Lindeque, P.K.; Rodriguez-Ezpeleta, N.; Albaina, A.; Lehtiniemi, M. Metabarcoding of Marine Zooplankton: Prospects, Progress and Pitfalls. J. Plankton Res. 2016, 38, 393–400. [Google Scholar] [CrossRef]
- Brown, E.A.; Chain, F.J.J.; Zhan, A.; MacIsaac, H.J.; Cristescu, M.E. Early Detection of Aquatic Invaders Using Metabarcoding Reveals a High Number of Non-Indigenous Species in Canadian Ports. Divers. Distrib. 2016, 22, 1045–1059. [Google Scholar] [CrossRef]
- Chain, F.J.J.; Brown, E.A.; Macisaac, H.J.; Cristescu, M.E. Metabarcoding Reveals Strong Spatial Structure and Temporal Turnover of Zooplankton Communities among Marine and Freshwater Ports. Divers. Distrib. 2016, 22, 493–504. [Google Scholar] [CrossRef]
- Santoferrara, L.F. Current Practice in Plankton Metabarcoding: Optimization and Error Management. J. Plankton Res. 2019, 41, 571–582. [Google Scholar] [CrossRef]
- Blanco-Bercial, L. Metabarcoding Analyses and Seasonality of the Zooplankton Community at BATS. Front. Mar. Sci. 2020, 7, 1–16. [Google Scholar] [CrossRef]
- Brandão, M.C.; Comtet, T.; Pouline, P.; Cailliau, C.; Blanchet-Aurigny, A.; Sourisseau, M.; Siano, R.; Memery, L.; Viard, F.; Nunes, F. Oceanographic Structure and Seasonal Variation Contribute to High Heterogeneity in Mesozooplankton over Small Spatial Scales. ICES J. Mar. Sci. 2021, 78, 3288–3302. [Google Scholar] [CrossRef]
- Lindeque, P.K.; Parry, H.E.; Harmer, R.A.; Somerfield, P.J.; Atkinson, A. Next Generation Sequencing Reveals the Hidden Diversity of Zooplankton Assemblages. PLoS ONE 2013, 8, e81327. [Google Scholar] [CrossRef] [PubMed]
- Abad, D.; Albaina, A.; Aguirre, M.; Laza-Martínez, A.; Uriarte, I.; Iriarte, A.; Villate, F.; Estonba, A. Is Metabarcoding Suitable for Estuarine Plankton Monitoring? A Comparative Study with Microscopy. Mar. Biol. 2016, 163, 1–13. [Google Scholar] [CrossRef]
- Harvey, J.B.J.; Johnson, S.B.; Fisher, J.L.; Peterson, W.T.; Vrijenhoek, R.C. Comparison of Morphological and next Generation DNA Sequencing Methods for Assessing Zooplankton Assemblages. J. Exp. Mar. Biol. Ecol. 2017, 487, 113–126. [Google Scholar] [CrossRef]
- Schroeder, A.; Stanković, D.; Pallavicini, A.; Gionechetti, F.; Pansera, M.; Camatti, E. DNA Metabarcoding and Morphological Analysis—Assessment of Zooplankton Biodiversity in Transitional Waters. Mar. Environ. Res. 2020, 160, 104946. [Google Scholar] [CrossRef]
- Coguiec, E.; Ershova, E.A.; Daase, M.; Vonnahme, T.R.; Wangensteen, O.S.; Gradinger, R.; Præbel, K.; Berge, J. Seasonal Variability in the Zooplankton Community Structure in a Sub-Arctic Fjord as Revealed by Morphological and Molecular Approaches. Front. Mar. Sci. 2021, 8, 705042. [Google Scholar] [CrossRef]
- Bucklin, A.; Batta-Lona, P.G.; Questel, J.M.; Wiebe, P.H.; Richardson, D.E.; Copley, N.J.; O’Brien, T.D. COI Metabarcoding of Zooplankton Species Diversity for Time-Series Monitoring of the NW Atlantic Continental Shelf. Front. Mar. Sci. 2022, 9, 867893. [Google Scholar] [CrossRef]
- Bucklin, A.; Yeh, H.D.; Questel, J.M.; Richardson, D.E.; Reese, B.; Copley, N.J.; Wiebe, P.H. Time-Series Metabarcoding Analysis of Zooplankton Diversity of the NW Atlantic Continental Shelf. ICES J. Mar. Sci. 2019, 76, 1162–1176. [Google Scholar] [CrossRef]
- Ershova, E.A.; Wangensteen, O.S.; Descoteaux, R.; Barth-Jensen, C.; Præbel, K. Metabarcoding as a Quantitative Tool for Estimating Biodiversity and Relative Biomass of Marine Zooplankton. ICES J. Mar. Sci. 2021, 78, 3342–3355. [Google Scholar] [CrossRef]
- Zheng, L.; He, J.; Lin, Y.; Cao, W.; Zhang, W. 16S RRNA Is a Better Choice than COI for DNA Barcoding Hydrozoans in the Coastal Waters of China. Acta Oecol. 2014, 33, 55–76. [Google Scholar] [CrossRef]
- Zhang, G.K.; Chain, F.J.J.; Abbott, C.L.; Cristescu, M.E. Metabarcoding Using Multiplexed Markers Increases Species Detection in Complex Zooplankton Communities. Evol. Appl. 2018, 11, 1901–1914. [Google Scholar] [CrossRef] [PubMed]
- Carroll, E.L.; Gallego, R.; Sewell, M.A.; Zeldis, J.; Ranjard, L.; Ross, H.A.; Tooman, L.K.; O’Rorke, R.; Newcomb, R.D.; Constantine, R. Multi-Locus DNA Metabarcoding of Zooplankton Communities and Scat Reveal Trophic Interactions of a Generalist Predator. Sci. Rep. 2019, 9, 281. [Google Scholar] [CrossRef] [PubMed]
- Ramos, S.; Cowen, R.K.; Ré, P.; Bordalo, A.A. Temporal and Spatial Distributions of Larval Fish Assemblages in the Lima Estuary (Portugal). Estuar. Coast. Shelf Sci. 2006, 66, 303–314. [Google Scholar] [CrossRef]
- Ramos, S.; Cowen, R.K.; Paris, C.; Ré, P.; Bordalo, A.A. Environmental Forcing and Larval Fish Assemblage Dynamics in the Lima River Estuary (Northwest Portugal). J. Plankton Res. 2006, 28, 275–286. [Google Scholar] [CrossRef]
- Sousa, R.; Dias, S.; Antunes, J.C. Spatial Subtidal Macrobenthic Distribution in Relation to Abiotic Conditions in the Lima Estuary, NW of Portugal. Hydrobiologia 2006, 559, 135–148. [Google Scholar] [CrossRef]
- Costa-Dias, S.; Sousa, R.; Antunes, C. Ecological Quality Assessment of the Lower Lima Estuary. Mar. Pollut. Bull. 2010, 61, 234–239. [Google Scholar] [CrossRef]
- Azevedo, I.; Ramos, S.; Mucha, A.P.; Bordalo, A.A. Applicability of Ecological Assessment Tools for Management Decision-Making: A Case Study from the Lima Estuary (NW Portugal). Ocean Coast. Manag. 2013, 72, 54–63. [Google Scholar] [CrossRef]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A New Versatile Primer Set Targeting a Short Fragment of the Mitochondrial COI Region for Metabarcoding Metazoan Diversity: Application for Characterizing Coral Reef Fish Gut Contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Lobo, J.; Costa, P.M.; Teixeira, M.A.L.; Ferreira, M.S.G.; Costa, M.H.; Costa, F.O. Enhanced Primers for Amplification of DNA Barcodes from a Broad Range of Marine Metazoans. BMC Ecol. 2013, 13, 34. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.W.; Richards, T.A. Multiple Marker Parallel Tag Environmental DNA Sequencing Reveals a Highly Complex Eukaryotic Community in Marine Anoxic Water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Leite, B.R.; Vieira, P.E.; Troncoso, J.S.; Costa, F.O. Comparing Species Detection Success between Molecular Markers in DNA Metabarcoding of Coastal Macroinvertebrates. Metabarcoding Metagenom. 2021, 5, 249–260. [Google Scholar] [CrossRef]
- Hollatz, C.; Leite, B.R.; Lobo, J.; Froufe, H.; Egas, C.; Costa, F.O. Priming of a DNA Metabarcoding Approach for Species Identification and Inventory in Marine Macrobenthic Communities. Genome 2017, 60, 260–271. [Google Scholar] [CrossRef] [PubMed]
- Clarke, L.J.; Beard, J.M.; Swadling, K.M.; Deagle, B.E. Effect of Marker Choice and Thermal Cycling Protocol on Zooplankton DNA Metabarcoding Studies. Ecol. Evol. 2017, 7, 873–883. [Google Scholar] [CrossRef]
- Pochon, X.; Bott, N.J.; Smith, K.F.; Wood, S.A. Evaluating Detection Limits of Next-Generation Sequencing for the Surveillance and Monitoring of International Marine Pests. PLoS ONE 2013, 8, e73935. [Google Scholar] [CrossRef]
- Zhan, A.; Hulák, M.; Sylvester, F.; Huang, X.; Adebayo, A.A.; Abbott, C.L.; Adamowicz, S.J.; Heath, D.D.; Cristescu, M.E.; Macisaac, H.J. High Sensitivity of 454 Pyrosequencing for Detection of Rare Species in Aquatic Communities. Methods Ecol. Evol. 2013, 4, 558–565. [Google Scholar] [CrossRef]
- van der Loos, L.M.; Nijland, R. Biases in Bulk: DNA Metabarcoding of Marine Communities and the Methodology Involved. Mol. Ecol. 2021, 30, 3270–3288. [Google Scholar] [CrossRef]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Schmieder, R.; Edwards, R. Quality Control and Preprocessing of Metagenomic Datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing Mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the Miseq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S. mBRAVE: The Multiplex Barcode Research And Visualization Environment. Biodivers. Inf. Sci. Stand. 2019, 3. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System: Barcoding. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Pruesse, E.; Peplies, J.; Glöckner, F.O. SINA: Accurate High-Throughput Multiple Sequence Alignment of Ribosomal RNA Genes. Bioinformatics 2012, 28, 1823–1829. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A Versatile Open Source Tool for Metagenomics. PeerJ 2016, 2016, e2584. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Duarte, S.; Vieira, P.E.; Leite, B.R.; Teixeira, M.A.L.; Neto, J.M.; Costa, F.O. Macrozoobenthos Monitoring in Portuguese Transitional Waters in the Scope of the Water Framework Directive Using Morphology and DNA Metabarcoding. Estuar. Coast. Shelf Sci. 2023, 281, 108207. [Google Scholar] [CrossRef]
- Fais, M.; Duarte, S.; Vieira, P.E.; Sousa, R.; Hajibabaei, M.; Canchaya, C.A.; Costa, F.O. Small-Scale Spatial Variation of Meiofaunal Communities in Lima Estuary (NW Portugal) Assessed through Metabarcoding. Estuar. Coast. Shelf Sci. 2020, 238, 106683. [Google Scholar] [CrossRef]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package - R Package Version 2.6-4. CRAN’s Repos. 2022. [Google Scholar]
- Tennekes, M.; Ellis, P. Treemap Visualization. CRAN’s Repos. 2022. [Google Scholar]
- Clarke, K.R.; Warwick, R.M. A Taxonomic Distinctness Index and Its Statistical Properties. J. Appl. Ecol. 1998, 35, 523–531. [Google Scholar] [CrossRef]
- Kolde, R. Pretty Heatmaps. CRAN’s Repos. 2022. [Google Scholar]
- Heberle, H.; Meirelles, G.V.; Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 7. [Google Scholar] [CrossRef]
- Radulovici, A.E.; Vieira, P.E.; Duarte, S.; Teixeira, M.A.L.; Borges, L.M.S.; Deagle, B.E.; Majaneva, S.; Redmond, N.; Schultz, J.A.; Costa, F.O. Revision and Annotation of DNA Barcode Records for Marine Invertebrates: Report of the 8th IBOL Conference Hackathon. Metabarcoding Metagenom. 2021, 5, 207–217. [Google Scholar] [CrossRef]
- Lobo, J.; Teixeira, M.A.L.; Borges, L.M.S.; Ferreira, M.S.G.; Hollatz, C.; Gomes, P.T.; Sousa, R.; Ravara, A.; Costa, M.H.; Costa, F.O. Starting a DNA Barcode Reference Library for Shallow Water Polychaetes from the Southern European Atlantic Coast. Mol. Ecol. Resour. 2016, 16, 298–313. [Google Scholar] [CrossRef]
- Lobo, J.; Ferreira, M.S.; Antunes, I.C.; Teixeira, M.A.L.; Borges, L.M.S.; Sousa, R.; Gomes, P.A.; Helena Costa, M.; Cunha, M.R.; Costa, F.O. Contrasting Morphological and DNA Barcode-Suggested Species Boundaries among Shallow-Water Amphipod Fauna from the Southern European Atlantic Coast. Genome 2017, 60, 147–157. [Google Scholar] [CrossRef]
- Leite, B.R.; Vieira, P.E.; Teixeira, M.A.L.; Lobo-Arteaga, J.; Hollatz, C.; Borges, L.M.S.; Duarte, S.; Troncoso, J.S.; Costa, F.O. Gap-Analysis and Annotated Reference Library for Supporting Macroinvertebrate Metabarcoding in Atlantic Iberia. Reg. Stud. Mar. Sci. 2020, 36, 101307. [Google Scholar] [CrossRef]
- Borges, L.M.S.; Hollatz, C.; Lobo, J.; Cunha, A.M.; Vilela, A.P.; Calado, G.; Coelho, R.; Costa, A.C.; Ferreira, M.S.G.; Costa, M.H.; et al. With a Little Help from DNA Barcoding: Investigating the Diversity of Gastropoda from the Portuguese Coast. Sci. Rep. 2016, 6, 20226. [Google Scholar] [CrossRef] [PubMed]
- Fontes, J.T.; Vieira, P.E.; Ekrem, T.; Soares, P.; Costa, F.O. BAGS: An Automated Barcode, Audit & Grade System for DNA Barcode Reference Libraries. Mol. Ecol. 2021, 21, 573–583. [Google Scholar] [CrossRef]
- Lavrador, A.S.; Fontes, J.T.; Vieira, P.E.; Costa, F.O.; Duarte, S. Compilation, Revision, and Annotation of DNA Barcodes of Marine Invertebrate Non-Indigenous Species (NIS) Occurring in European Coastal Regions. Diversity 2023, 15, 174. [Google Scholar] [CrossRef]
- Oliveira, L.M.; Knebelsberger, T.; Landi, M.; Soares, P.; Raupach, M.J.; Costa, F.O. Assembling and Auditing a Comprehensive DNA Barcode Reference Library for European Marine Fishes. J. Fish Biol. 2016, 89, 2741–2754. [Google Scholar] [CrossRef] [PubMed]
- Moutinho, J. DNA Metabarcoding Monitoring of Zooplankton for the Detection of Non-Indigenous Species (NIS): A Seasonal Study in a Recreational Marina of the Northwest of Portugal. Master’s Thesis, University of Minho, Braga, Portugal, 2022. [Google Scholar]
- Ramos, S.; Cabral, H.; Elliott, M. Do Fish Larvae Have Advantages over Adults and Other Components for Assessing Estuarine Ecological Quality? Ecol. Indic. 2015, 55, 74–85. [Google Scholar] [CrossRef]
- Ramos, S.; Ré, P.; Bordalo, A.A. Recruitment of Flatfish Species to an Estuarine Nursery Habitat (Lima Estuary, NW Iberian Peninsula). J. Sea Res. 2010, 64, 473–486. [Google Scholar] [CrossRef]
- Ramos, S. Ichthyoplankton of the Lima Estuary (NW Portugal): Ecology of the Early Life Stages of Pleuronectiformes; University of Porto: Porto, Portugal, 2007. [Google Scholar]
- Guimarães, C.; Galhano, H. Ecological Study of the Estuary of River Lima (Portugal): I—The North Bank Saltmarshes. In Publicações do Instituito de Zoologia “Dr. Augusto Nobre”; Faculdade de Ciências do Porto: Porto, Portugal, 1987; pp. 1–54. [Google Scholar]
- Guimarães, C.; Galhano, H. Ecological Study of the Estuary of River Lima (Portugal): II—A Mud-Sandybeach. In Publicações do Instituito de Zoologia “Dr. Augusto Nobre”; Faculdade de Ciências do Porto: Porto, Portugal, 1988; pp. 1–73. [Google Scholar]
- Guimarães, C.; Galhano, H. Ecological Study of the Estuary of River Lima (Portugal): III—Channels of Darque. In Publicações do Instituito de Zoologia “Dr. Augusto Nobre”; Faculdade de Ciências do Porto: Porto, Portugal, 1989; pp. 1–52. [Google Scholar]
- Stefanni, S.; Stanković, D.; Borme, D.; de Olazabal, A.; Juretić, T.; Pallavicini, A.; Tirelli, V. Multi-Marker Metabarcoding Approach to Study Mesozooplankton at Basin Scale. Sci. Rep. 2018, 8, 12085. [Google Scholar] [CrossRef]
- Belmonte, G.; Rubino, F. Resting Cysts from Coastal Marine Plankton. In Oceanography and Marine Biology an Annual Review; Hawkins, S.J., Allcock, A.L., Bates, A.E., Firth, L.B., Smith, I.P., Swearer, S.E., Todd, P.A., Eds.; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Milton Park, UK, 2019; Volume 57, pp. 1–88. [Google Scholar]
- Belmonte, G.; Vaglio, I.; Rubino, F.; Alabiso, G. Zooplankton Composition along the Confinement Gradient of the Taranto Sea System (Ionian Sea, South-Eastern Italy). J. Mar. Syst. 2013, 128, 222–238. [Google Scholar] [CrossRef]
- Rubino, F.; Belmonte, G. Habitat Shift for Plankton: The Living Side of Benthic-Pelagic Coupling in the Mar Piccolo of Taranto (Southern Italy, Ionian Sea). Water 2021, 13, 3619. [Google Scholar] [CrossRef]
- Intxausti, L.; Villate, F.; Uriarte, I.; Iriarte, A.; Ameztoy, I. Size-Related Response of Zooplankton to Hydroclimatic Variability and Water-Quality in an Organically Polluted Estuary of the Basque Coast (Bay of Biscay). J. Mar. Syst. 2012, 94, 87–96. [Google Scholar] [CrossRef]
- Sousa, R. Estrutura Das Comunidades de Macroinvertebrados Bentónicos Presentes No Estuário Do Rio Lima; University of Porto: Porto, Portugal, 2003. [Google Scholar]
- Largier, J.; Delgadillo, F.; Grierson, P. Seasonally Hypersaline Estuaries in Mediterranean-Climate Regions. Estuar. Coast. Shelf Sci. 1997, 45, 789–797. [Google Scholar] [CrossRef]
- Valente, A.C.N.; Alexandrino, P.J.B. Ecological Study of the Estuary of River Lima. IV. The Ichthyofauna in the Darque Channels (River Lima Estuary) with Special Reference to the Biology of the Sand-Melt, Atherina presbyter Cuvier, 1829 (Pisces: Atherinidae). Publicações do Instituto de Zoologia “Dr. Augusto Nobre” 1988, 202, 1–17. [Google Scholar]
- Hebert, P.D.N.; Ratnasingham, S.; DeWaard, J.R. Barcoding Animal Life: Cytochrome c Oxidase Subunit 1 Divergences among Closely Related Species. Proc. R. Soc. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Capra, E.; Giannico, R.; Montagna, M.; Turri, F.; Cremonesi, P.; Strozzi, F.; Leone, P.; Gandini, G.; Pizzi, F. A New Primer Set for DNA Metabarcoding of Soil Metazoa. Eur. J. Soil Biol. 2016, 77, 53–59. [Google Scholar] [CrossRef]
- Mueller, R.L. Evolutionary Rates, Divergence Dates, and the Performance of Mitochondrial Genes in Bayesian Phylogenetic Analysis. Syst. Biol. 2006, 55, 289–300. [Google Scholar] [CrossRef]
- Gouy, M.; Li, W.H. Molecular Phylogeny of the Kingdoms Animalia, Plantae, and Fungi. Mol. Biol. Evol. 1989, 6, 109–122. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A Method for Studying Protistan Diversity Using Massively Parallel Sequencing of V9 Hypervariable Regions of Small-Subunit Ribosomal RNA Genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Tang, C.Q.; Leasi, F.; Obertegger, U.; Kieneke, A.; Barraclough, T.G.; Fontaneto, D. The Widely Used Small Subunit 18S RDNA Molecule Greatly Underestimates True Diversity in Biodiversity Surveys of the Meiofauna. Proc. Natl. Acad. Sci. USA 2012, 109, 16208–16212. [Google Scholar] [CrossRef]
- Questel, J.M.; Hopcroft, R.R.; DeHart, H.M.; Smoot, C.A.; Kosobokova, K.N.; Bucklin, A. Metabarcoding of Zooplankton Diversity within the Chukchi Borderland, Arctic Ocean: Improved Resolution from Multi-Gene Markers and Region-Specific DNA Databases. Mar. Biodivers. 2021, 51, 1–19. [Google Scholar] [CrossRef]
- Moutinho, J.; Lavrador, A.S.; Vieira, P.E.; Costa, F.O.; Duarte, S. Assessing the Seasonal Dynamics of Zooplankton in a Recreational Marina of the Northwest of Portugal through Multi-Marker DNA Metabarcoding. In Proceedings of the ARPHA Conference Abstracts; Pensoft Publishers: Sofia, Bulgaria, 2022; Volume 5. [Google Scholar]
- Lavrador, A.S.; Amaral, F.G.; Moutinho, J.; Vieira, P.E.; Costa, F.O.; Duarte, S. Detection and Monitoring of Non-Indigenous Invertebrate Species in Recreational Marinas through DNA Metabarcoding of Zooplankton Communities in the North of Portugal. In Proceedings of the MetaZooGene Symposium: New Insights into Biodiversity, Biogeography, Ecology, and Evolution of Marine Zooplankton Based on Molecular Approaches, Dublin, Ireland, 23 September 2022; Available online: https://metazoogene.org/symposium2022 (accessed on 12 December 2023).
- Lavrador, A.; Amaral, F.; Vieira, P.E.; Costa, F.; Duarte, S. Surveillance of Non-Indigenous Invertebrate Species through DNA Metabarcoding in Recreational Marinas in the North and Center of Portugal. In Proceedings of the ARPHA Conference Abstracts; Pensoft Publishers: Sofia, Bulgaria, 2021; Volume 4. [Google Scholar]
- Katsanevakis, S.; Bogucarskis, K.; Gatto, F.; Vandekerkhove, J.; Deriu, I.; Cardoso, A.C. Building the European Alien Species Information Network (EASIN): A Novel Approach for the Exploration of Distributed Alien Species Data. Bioinvasions Rec. 2012, 1, 235–245. [Google Scholar] [CrossRef]
- Luiz, O.J.; Comin, E.J.; Madin, J.S. Far Away from Home: The Occurrence of the Indo-Pacific Bannerfish Heniochus Acuminatus (Pisces: Chaetodontidae) in the Atlantic. Bull. Mar. Sci. 2014, 90, 741–744. [Google Scholar] [CrossRef]
- Adelir-Alves, J.; Soeth, M.; Braga, R.R.; Spach, H.L. Non-Native Reef Fishes in the Southwest Atlantic Ocean: A Recent Record of Heniochus Acuminatus (Linnaeus, 1758) (Perciformes, Chaetodontidae) and Biological Aspects of Chromis Limbata (Valenciennes, 1833) (Perciformes, Pomacentridae). Check List 2018, 14, 379–385. [Google Scholar] [CrossRef]
- Png-Gonzalez, L.; Comas-González, R.; Calvo-Manazza, M.; Follana-Berná, G.; Ballesteros, E.; Díaz-Tapia, P.; Falcón, J.M.; García Raso, J.E.; Gofas, S.; González-Porto, M.; et al. Updating the National Baseline of Non-Indigenous Species in Spanish Marine Waters. Diversity 2023, 15, 630. [Google Scholar] [CrossRef]
- Obst, M. 18S Metabarcoding Genetic Observations of Marine Species in the Port of Wallhamn, Sweden (2022); University of Gothenburg: Göteborg, Sweden, 2023. [Google Scholar]
- Hoeh, W.R.; Blakley, K.H.; Brown, W.M. Heteroplasmy Suggests Limited Biparental Inheritance of Mytilus Mitochondrial DNA. Science (1979) 1991, 251, 1488–1490. [Google Scholar] [CrossRef] [PubMed]
- Śmietanka, B.; Burzyński, A.; Hummel, H.; Wenne, R. Glacial History of the European Marine Mussels Mytilus, Inferred from Distribution of Mitochondrial DNA Lineages. Heredity 2014, 113, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Boukadida, K.; Mlouka, R.; Clerandeau, C.; Banni, M.; Cachot, J. Natural Distribution of Pure and Hybrid Mytilus Sp. along the South Mediterranean and North-East Atlantic Coasts and Sensitivity of D-Larvae Stages to Temperature Increases and Metal Pollution. Sci. Total Environ. 2021, 756, 143675. [Google Scholar] [CrossRef]
- Hilbish, T.J.; Mullinax, A.; Dolven, S.I.; Meyer, A.; Koehn, R.K.; Rawson, P.D. Origin of the Antitropical Distribution Pattern in Marine Mussels (Mytilus Spp.): Routes and Timing of Transequatorial Migration. Mar. Biol. 2000, 136, 69–77. [Google Scholar] [CrossRef]
- Wangensteen, O.S.; Palacín, C.; Guardiola, M.; Turon, X. DNA Metabarcoding of Littoral Hardbottom Communities: High Diversity and Database Gaps Revealed by Two Molecular Markers. PeerJ 2018, 2018, e4705. [Google Scholar] [CrossRef]
- Chainho, P.; Fernandes, A.; Amorim, A.; Ávila, S.P.; Canning-Clode, J.; Castro, J.J.; Costa, A.C.; Costa, J.L.; Cruz, T.; Gollasch, S.; et al. Non-Indigenous Species in Portuguese Coastal Areas, Coastal Lagoons, Estuaries and Islands. Estuar. Coast. Shelf Sci. 2015, 167, 199–211. [Google Scholar] [CrossRef]
- Marques, S.C.; Pardal, M.A.; Pereira, M.J.; Gonçalves, F.; Marques, J.C.; Azeiteiro, U.M. Zooplankton Distribution and Dynamics in a Temperate Shallow Estuary. Hydrobiologia 2007, 587, 213–223. [Google Scholar] [CrossRef]
- Morais, P.; Chícharo, M.A.; Chícharo, L. Changes in a Temperate Estuary during the Filling of the Biggest European Dam. Sci. Total Environ. 2009, 407, 2245–2259. [Google Scholar] [CrossRef] [PubMed]
- Illumina. 16S Metagenomic Sequencing Library Preparation Preparing 16S Ribosomal RNA Gene Amplicons for the Illumina MiSeq System; Illumina Technical Document, (Part. No. 15044223 Rev. B.); Illumina: Minato, Tokyo, 2013. [Google Scholar]
- Lejzerowicz, F.; Esling, P.; Pillet, L.; Wilding, T.A.; Black, K.D.; Pawlowski, J. High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 2015, 5, 13932. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging Findings Background. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]







| Taxonomic Group | Season | Site | ||
|---|---|---|---|---|
| F | p | F | p | |
| Zooplankton | 9.57 | <0.01 | 2.54 | <0.01 |
| Mollusca | 10.91 | <0.01 | 1.91 | <0.01 |
| Arthropoda | 7.26 | <0.01 | 3.97 | <0.01 |
| Annelida | 11.18 | <0.01 | 2.08 | <0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moutinho, J.; Carreira-Flores, D.; Gomes, P.T.; Costa, F.O.; Duarte, S. Assessing the Seasonal and Spatial Dynamics of Zooplankton through DNA Metabarcoding in a Temperate Estuary. Animals 2023, 13, 3876. https://doi.org/10.3390/ani13243876
Moutinho J, Carreira-Flores D, Gomes PT, Costa FO, Duarte S. Assessing the Seasonal and Spatial Dynamics of Zooplankton through DNA Metabarcoding in a Temperate Estuary. Animals. 2023; 13(24):3876. https://doi.org/10.3390/ani13243876
Chicago/Turabian StyleMoutinho, Jorge, Diego Carreira-Flores, Pedro T. Gomes, Filipe O. Costa, and Sofia Duarte. 2023. "Assessing the Seasonal and Spatial Dynamics of Zooplankton through DNA Metabarcoding in a Temperate Estuary" Animals 13, no. 24: 3876. https://doi.org/10.3390/ani13243876
APA StyleMoutinho, J., Carreira-Flores, D., Gomes, P. T., Costa, F. O., & Duarte, S. (2023). Assessing the Seasonal and Spatial Dynamics of Zooplankton through DNA Metabarcoding in a Temperate Estuary. Animals, 13(24), 3876. https://doi.org/10.3390/ani13243876









