Development of a Multiplex Crystal Digital RT-PCR for Differential Detection of Classical, Highly Pathogenic, and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral Strains
2.2. Clinical Samples
2.3. Design of Primers and Probes
2.4. Extraction of Nucleic Acids
2.5. Construction of the Standard Plasmids
2.6. Optimization of the Reaction Conditions
2.7. Specificity Analysis
2.8. Sensitivity Analysis
2.9. Repeatability Analysis
2.10. Detection of the Clinical Samples
3. Results
3.1. Construction of the Standard Plasmids
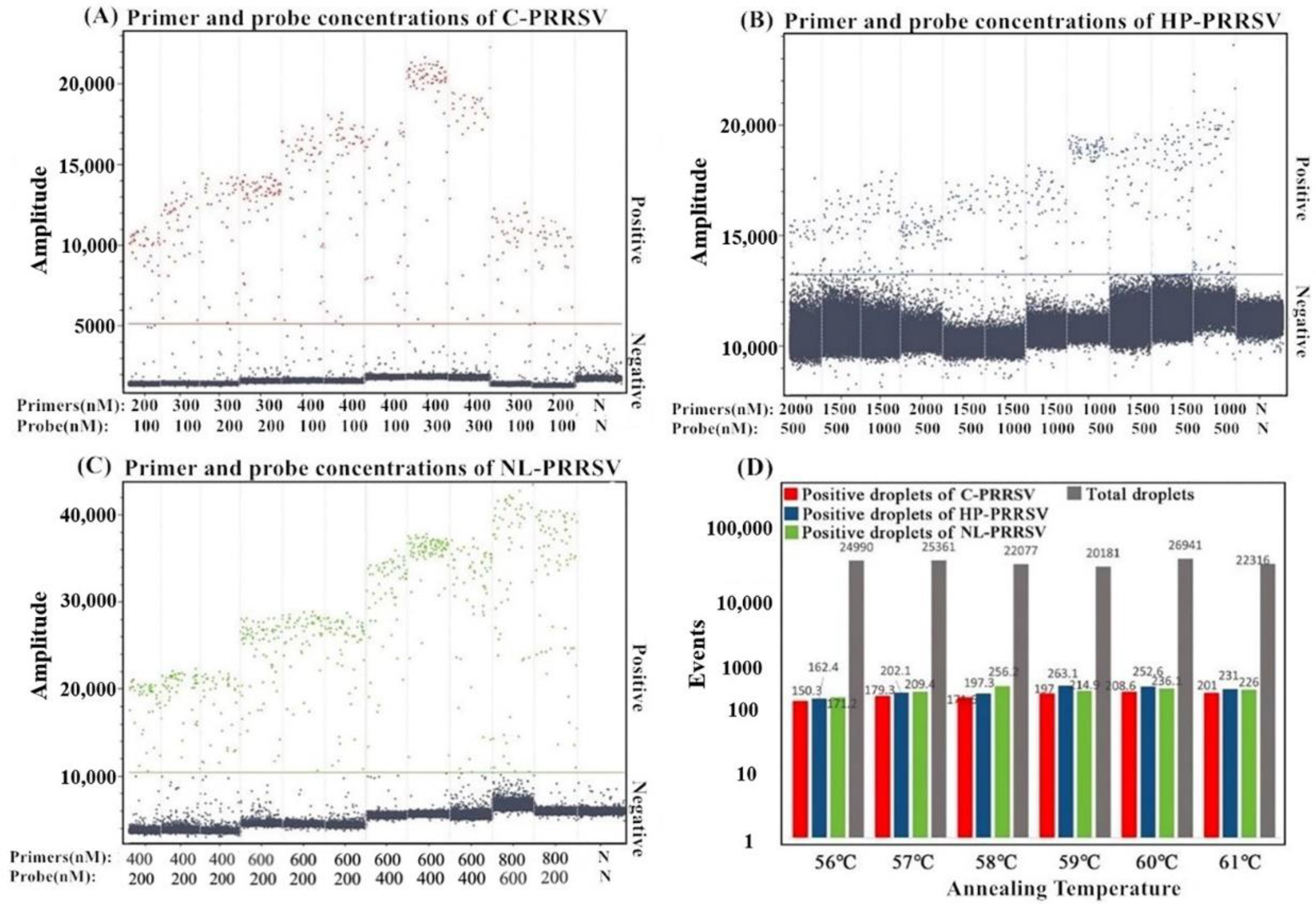
3.2. Determination of the Optimal Reaction Conditions
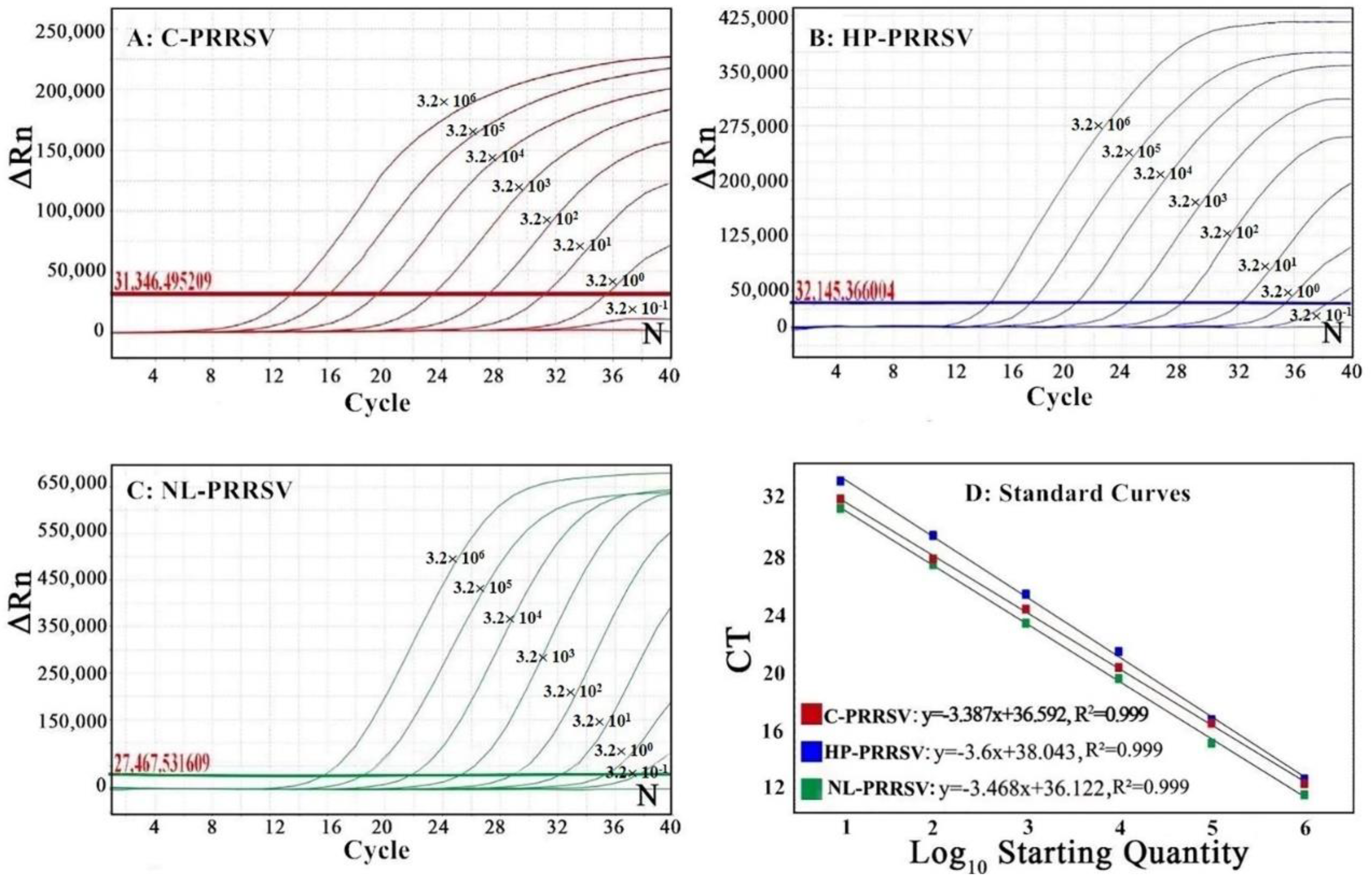
3.3. Generation of the Standard Curves
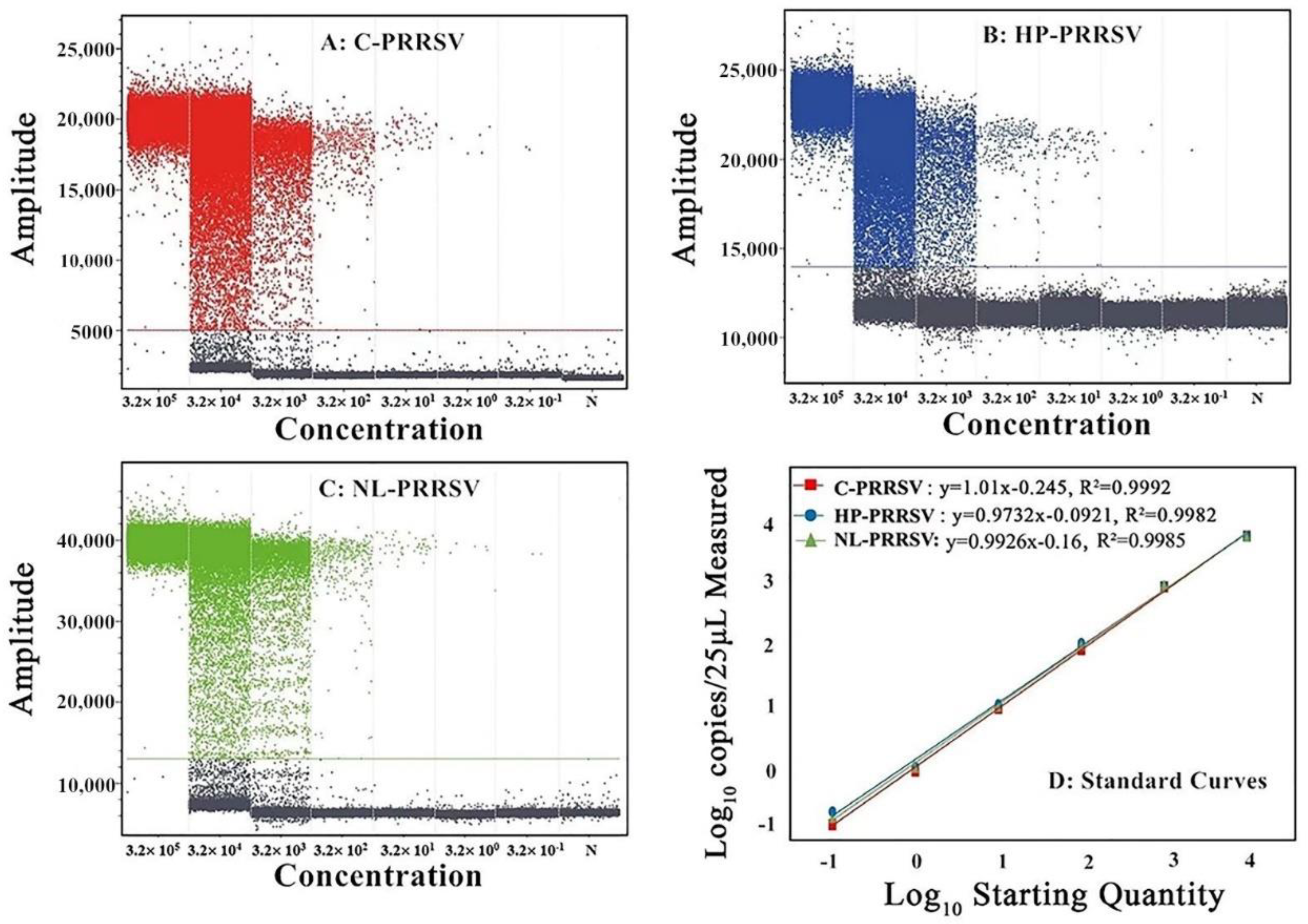
3.4. Sensitivity Analysis
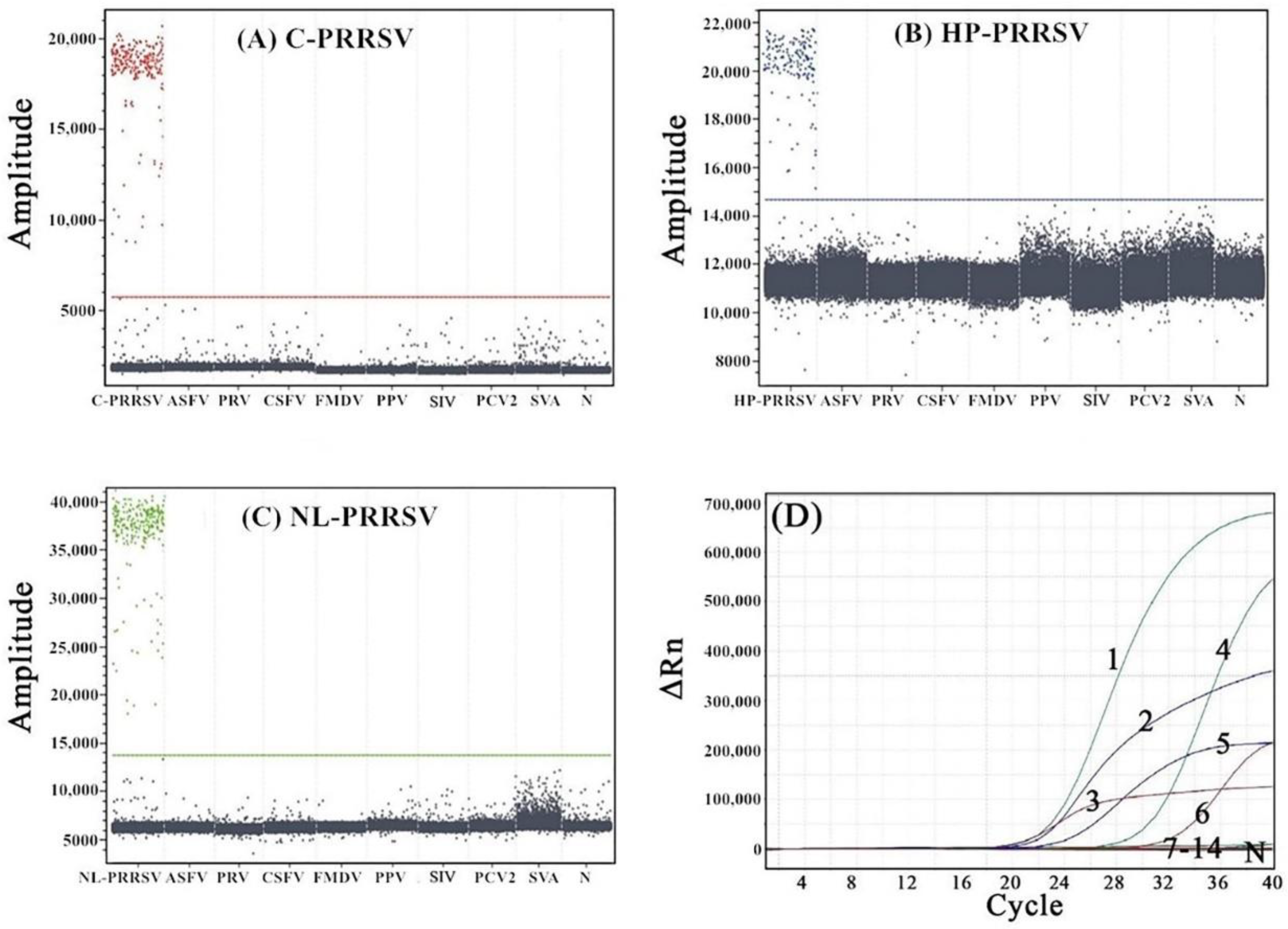
3.5. Specificity Analysis
3.6. Repeatability Analysis
3.7. Application for Detection of Clinical Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lunney, J.K.; Benfield, D.A.; Rowland, R.R. Porcine reproductive and respiratory syndrome virus: An update on an emerging and re-emerging viral disease of swine. Virus Res. 2010, 154, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Kappes, M.A.; Faaberg, K.S. PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479–480, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Montaner-Tarbes, S.; Del Portillo, H.A.; Montoya, M.; Fraile, L. Key gaps in the knowledge of the porcine respiratory reproductive syndrome virus (PRRSV). Front. Vet. Sci. 2019, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Rossow, K.D. Porcine reproductive and respiratory syndrome. Vet. Pathol. 1998, 35, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Yang, H. Porcine reproductive and respiratory syndrome in China. Virus Res. 2010, 154, 31–37. [Google Scholar] [CrossRef]
- Tian, K.; Yu, X.; Zhao, T.; Feng, Y.; Cao, Z.; Wang, C.; Hu, Y.; Chen, X.; Hu, D.; Tian, X.; et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Loving, C.L.; Vorwald, A.C.; Kehrli, M.E., Jr.; Baker, R.B.; Nicholson, T.L.; Lager, K.M.; Miller, L.C.; Faaberg, K.S. Genomic sequence and virulence comparison of four type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 2012, 169, 212–221. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Z.; Ding, Y.; Ge, X.; Guo, X.; Yang, H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 2015, 21, 2256–2257. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, X.X.; Li, R.; Qiao, S.; Zhang, G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: A molecular epidemiological perspective. Virol. J. 2018, 15, 2. [Google Scholar] [CrossRef]
- Chen, N.; Xiao, Y.; Ye, M.; Li, X.; Li, S.; Xie, N.; Wei, Y.; Wang, J.; Zhu, J. High genetic diversity of Chinese porcine reproductive and respiratory syndrome viruses from 2016 to 2019. Res. Vet. Sci. 2020, 131, 38–42. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Xiao, S.; Yang, X.; Chen, X.; Wu, P.; Song, J.; Ma, Z.; Cai, Z.; Jiang, M.; et al. High-frequency mutation and recombination are responsible for the emergence of novel porcine reproductive and respiratory syndrome virus in northwest China. Arch. Virol. 2019, 164, 2725–2733. [Google Scholar] [CrossRef]
- Zhao, H.Z.; Wang, F.X.; Han, X.Y.; Guo, H.; Liu, C.Y.; Hou, L.N.; Wang, Y.X.; Zheng, H.; Wang, L.; Wen, Y.J. Recent advances in the study of NADC34-like porcine reproductive and respiratory syndrome virus in China. Front. Microbiol. 2022, 13, 950402. [Google Scholar] [CrossRef] [PubMed]
- Tian, K. NADC30-like porcine reproductive and respiratory syndrome in China. Open Virol. J. 2017, 11, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Q.; Cao, Z.; Tang, Y.D.; Xia, D.; Wang, G.; Shan, H. Recent advances in porcine reproductive and respiratory syndrome virus NADC30-like research in China: Molecular characterization, pathogenicity, and control. Front. Microbiol. 2022, 12, 791313. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, G.; Yu, L.; Li, L.; Zhang, Y.; Zhou, Y.; Tong, W.; Liu, C.; Gao, F.; Tong, G. Genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV) from 1996 to 2017 in China. Front. Microbiol. 2020, 11, 618. [Google Scholar] [CrossRef]
- Fang, K.; Liu, S.; Li, X.; Chen, H.; Qian, P. Epidemiological and genetic characteristics of porcine reproductive and respiratory syndrome virus in South China between 2017 and 2021. Front. Vet. Sci. 2022, 9, 853044. [Google Scholar] [CrossRef] [PubMed]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R.; et al. Real-time PCR in clinical microbiology: Applications for routine laboratory testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef] [PubMed]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Kojabad, A.A.; Farzanehpour, M.; Galeh, H.E.G.; Dorostkar, R.; Jafarpour, A.; Bolandian, M.; Nodooshan, M.M. Droplet digital PCR of viral DNA/RNA, current progress, challenges, and future perspectives. J. Med. Virol. 2021, 93, 4182–4197. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Kuypers, J.; Jerome, K.R. Applications of digital PCR for clinical microbiology. J. Clin. Microbiol. 2017, 55, 1621–1628. [Google Scholar] [CrossRef] [PubMed]
- Salipante, S.J.; Jerome, K.R. Digital PCR-An emerging technology with broad applications in microbiology. Clin. Chem. 2020, 66, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Egli, C.; Thür, B.; Liu, L.; Hofmann, M.A. Quantitative TaqMan RT-PCR for the detection and differentiation of European and North American strains of porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2001, 98, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Kleiboeker, S.B.; Schommer, S.K.; Lee, S.M.; Watkins, S.; Chittick, W.; Polson, D. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase-PCR. J. Vet. Diagn. Investig. 2005, 17, 165–170. [Google Scholar] [CrossRef]
- Wernike, K.; Hoffmann, B.; Dauber, M.; Lange, E.; Schirrmeier, H.; Beer, M. Detection and typing of highly pathogenic porcine reproductive and respiratory syndrome virus by multiplex real-time RT-PCR. PLoS ONE 2012, 7, e38251. [Google Scholar] [CrossRef]
- Chen, N.; Ye, M.; Xiao, Y.; Li, S.; Huang, Y.; Li, X.; Tian, K.; Zhu, J. Development of universal and quadruplex real-time RT-PCR assays for simultaneous detection and differentiation of porcine reproductive and respiratory syndrome viruses. Transbound. Emerg. Dis. 2019, 66, 2271–2278. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Meng, K.; Liu, Y.; Zhang, Y.; Wang, Z.; Chen, Z.; Yang, J.; Sun, W.; Guo, L.; Ren, S.; et al. Simultaneous detection of classical PRRSV, highly pathogenic PRRSV and NADC30-like PRRSV by TaqMan probe real-time PCR. J. Virol. Methods 2019, 282, 113774. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Chen, Y.; Wang, L.; Gao, J.; Mo, D.; He, Z.; Liu, X. Simultaneous detection and differentiation of highly virulent and classical Chinese-type isolation of PRRSV by real-time RT-PCR. J. Immunol. Res. 2014, 2014, 809656. [Google Scholar] [CrossRef]
- Chen, N.H.; Chen, X.Z.; Hu, D.M.; Yu, X.L.; Wang, L.L.; Han, W.; Wu, J.J.; Cao, Z.; Wang, C.B.; Zhang, Q.; et al. Rapid differential detection of classical and highly pathogenic North American porcine reproductive and respiratory syndrome virus in China by a duplex real-time RT-PCR. J. Virol. Methods 2009, 161, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Chai, Z.; Ma, W.; Fu, F.; Lang, Y.; Wang, W.; Tong, G.; Liu, Q.; Cai, X.; Li, X. A SYBR Green-based real-time RT-PCR assay for simple and rapid detection and differentiation of highly pathogenic and classical type 2 porcine reproductive and respiratory syndrome virus circulating in China. Arch. Virol. 2013, 158, 407–415. [Google Scholar] [CrossRef]
- Yang, Q.; Xi, J.; Chen, X.; Hu, S.; Chen, N.; Qiao, S.; Wan, S.; Bao, D. The development of a sensitive droplet digital PCR for quantitative detection of porcine reproductive and respiratory syndrome virus. Int. J. Biol. Macromol. 2017, 104, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Shi, K.; Chen, Y.; Yin, Y.; Long, F.; Feng, S.; Liu, H.; Qu, S.; Si, H. A multiplex crystal digital PCR for detection of African swine fever virus, classical swine fever virus, and porcine reproductive and respiratory syndrome virus. Front. Vet. Sci. 2022, 9, 926881. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.L.; Loganathan, N.; Agarwalla, S.; Yang, C.; Yuan, W.; Zeng, J.; Wu, R.; Wang, W.; Duraiswamy, S. Current commercial dPCR platforms: Technology and market review. Crit. Rev. Biotechnol. 2022, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Madic, J.; Zocevic, A.; Senlis, V.; Fradet, E.; Andre, B.; Muller, S.; Dangla, R.; Droniou, M.E. Three-color crystal digital PCR. Biomol. Detect. Quantif. 2016, 10, 34–46. [Google Scholar] [CrossRef]
- Whale, A.S.; Cowen, S.; Foy, C.A.; Huggett, J.F. Methods for applying accurate digital PCR analysis on low copy DNA samples. PLoS ONE 2013, 8, e58177. [Google Scholar] [CrossRef]
- Whale, A.S.; Huggett, J.F.; Tzonev, S. Fundamentals of multiplexing with digital PCR. Biomol. Detect. Quantif. 2016, 10, 15–23. [Google Scholar] [CrossRef]
- Zhao, K.; Ye, C.; Chang, X.B.; Jiang, C.G.; Wang, S.J.; Cai, X.H.; Tong, G.Z.; Tian, Z.J.; Shi, M.; An, T.Q. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef]
- Zhang, Z.; Qu, X.; Zhang, H.; Tang, X.; Bian, T.; Sun, Y.; Zhou, M.; Ren, F.; Wu, P. Evolutionary and recombination analysis of porcine reproductive and respiratory syndrome isolates in China. Virus Genes 2020, 56, 354–360. [Google Scholar] [CrossRef]
- Cui, X.; Xia, D.; Huang, X.; Sun, Y.; Shi, M.; Zhang, J.; Li, G.; Yang, Y.; Wang, H.; Cai, X.; et al. Analysis of recombinant characteristics based on 949 PRRSV-2 genomic sequences obtained from 1991 to 2021 shows that viral multiplication ability contributes to dominant recombination. Microbiol. Spectr. 2022, 10, e0293422. [Google Scholar] [CrossRef]
- Li, Y.; Ji, G.; Xu, X.; Wang, J.; Li, Y.; Tan, F.; Li, X. Development and application of an RT-PCR to differentiate the prevalent NA-PRRSV strains in China. Open Virol. J. 2017, 11, 66–72. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Wang, J.; Bai, X.; Ji, G.; Yan, H.; Li, Y.; Wang, Y.; Tan, F.; Xiao, Y.; Li, X.; et al. Pathogenicity comparison between highly pathogenic and NADC30-like porcine reproductive and respiratory syndrome virus. Arch. Virol. 2016, 161, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, J.; Sun, Y.; Liu, X.; Gao, Y.; Wang, X.; Yang, Y.; Jiang, P. Comparison of pathogenicity of different subgenotype porcine reproductive and respiratory syndrome viruses isolated in China. Microb. Pathog. 2022, 168, 105607. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Zhao, T.; Peng, Z.; Sun, Y.; Stratton, C.W.; Zhou, D.; Tang, X.; Tian, Y.; Chen, H.; Wu, B. Epidemiological and genetic characteristics of porcine reproductive and respiratory syndrome virus circulating in central and south China in 2016. Acta Trop. 2019, 190, 83–91. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, Y.; Xia, Q.; Guan, Z.; Zhang, J.; Li, B.; Qiu, Y.; Liu, K.; Shao, D.; Ma, Z.; et al. Genetic characterization of porcine reproductive and respiratory syndrome virus from Eastern China during 2017–2022. Front. Microbiol. 2022, 13, 971817. [Google Scholar] [CrossRef]
- Chung, W.B.; Lin, M.W.; Chang, W.F.; Hsu, M.; Yang, P.C. Persistence of porcine reproductive and respiratory syndrome virus in intensive farrow-to-finish pig herds. Can. J. Vet. Res. 1997, 61, 292–298. [Google Scholar] [PubMed]
- Wills, R.W.; Doster, A.R.; Galeota, J.A.; Sur, J.H.; Osorio, F.A. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J. Clin. Microbiol. 2003, 41, 58–62. [Google Scholar] [CrossRef]
- Wang, G.; Song, T.; Yu, Y.; Liu, Y.; Shi, W.; Wang, S.; Rong, F.; Dong, J.; Liu, H.; Cai, X.; et al. Immune responses in piglets infected with highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2011, 142, 170–178. [Google Scholar] [CrossRef]
- Dwivedi, V.; Manickam, C.; Binjawadagi, B.; Linhares, D.; Murtaugh, M.P.; Renukaradhya, G.J. Evaluation of immune responses to porcine reproductive and respiratory syndrome virus in pigs during early stage of infection under farm conditions. Virol. J. 2012, 9, 45. [Google Scholar] [CrossRef]
- Loving, C.L.; Osorio, F.A.; Murtaugh, M.P.; Zuckermann, F.A. Innate and adaptive immunity against porcine reproductive and respiratory syndrome virus. Vet. Immunol. Immunopathol. 2015, 167, 1–14. [Google Scholar] [CrossRef]
- Wang, T.Y.; Sun, M.X.; Zhang, H.L.; Wang, G.; Zhan, G.; Tian, Z.J.; Cai, X.H.; Su, C.; Tang, Y.D. Evasion of antiviral innate immunity by porcine reproductive and respiratory syndrome virus. Front. Microbiol. 2021, 12, 693799. [Google Scholar] [CrossRef]
- Chen, N.; Huang, Y.; Ye, M.; Li, S.; Xiao, Y.; Cui, B.; Zhu, J. Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV) and porcine circoviruses (PCV2 and PCV3) in eight regions of China from 2016 to 2018. Infect. Genet. Evol. 2019, 68, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Fan, P.; Wei, Y.; Guo, L.; Wu, H.; Huang, L.; Liu, J.; Liu, C. Synergistic effects of sequential infection with highly pathogenic porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Virol. J. 2013, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Yang, B.; Yuan, X.; Shen, C.; Zhang, D.; Shi, X.; Zhang, T.; Cui, H.; Yang, J.; Chen, X.; et al. Advanced research in porcine reproductive and respiratory syndrome virus co-infection with other pathogens in swine. Front. Vet. Sci. 2021, 8, 699561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, P.; Xie, C.; Ha, Z.; Shi, N.; Zhang, H.; Li, Z.; Han, J.; Xie, Y.; Qiu, X.; et al. Synergistic pathogenicity by coinfection and sequential infection with NADC30-like PRRSV and PCV2 in post-weaned pigs. Viruses 2022, 14, 193. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, B.; Xu, L.; Jin, H.; Ge, X.; Guo, X.; Han, J.; Yang, H. Efficacy evaluation of three modified-live virus vaccines against a strain of porcine reproductive and respiratory syndrome virus NADC30-like. Vet. Microbiol. 2017, 207, 108–116. [Google Scholar] [CrossRef]
- Huang, Y.; Li, Z.; Li, J.; Kong, Y.; Yang, L.; Mah, C.K.; Liu, G.; Yu, B.; Wang, K. Efficacy evaluation of three modified-live PRRS vaccines against a local strain of highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2019, 229, 117–123. [Google Scholar] [CrossRef]
- Chai, W.; Liu, Z.; Sun, Z.; Su, L.; Zhang, C.; Huang, L. Efficacy of two porcine reproductive and respiratory syndrome (PRRS) modified-live virus (MLV) vaccines against heterologous NADC30-like PRRS virus challenge. Vet. Microbiol. 2020, 248, 108805. [Google Scholar] [CrossRef]
- Proctor, J.; Wolf, I.; Brodsky, D.; Cortes, L.M.; Frias-De-Diego, A.; Almond, G.W.; Crisci, E.; Negrão Watanabe, T.T.; Hammer, J.M.; Käser, T. Heterologous vaccine immunogenicity, efficacy, and immune correlates of protection of a modified-live virus porcine reproductive and respiratory syndrome virus vaccine. Front. Microbiol. 2022, 13, 977796. [Google Scholar] [CrossRef]
| Primer | Sequence (5′→3′) | Product Size (bp) |
|---|---|---|
| C-PRRSV-F | AGTTGGGAAGATTTGGCTGTTA | 234 |
| C-PRRSV-R | ACCTGCTGAAACTTACGCCGCG | |
| C-PRRSV-P | CY5-TCACCGCAATGCATCTTCAGGC-BHQ2 | |
| HP-PRRSV-F | GTCGCGACGTGTCCCCAAGCT | 172 |
| HP-PRRSV-R | GCCCATGTTCTGCGATGGT | |
| HP-PRRSV-P | FAM-CACCAGTTCCTGCACCGCGTAGAACT-BHQ1 | |
| NL-PRRSV-F | AACGTATTGGACACCTCTTTTG | 217 |
| NL-PRRSV-R | TGGACCTAATCTTCCTGCGTGGG | |
| NL-PRRSV-P | VIC-CGGTATTCCAGTCTCGAAAAGC-BHQ1 |
| Multiplex cdRT-PCR | Multiplex qRT-PCR | |||
|---|---|---|---|---|
| Volume (μL) | Final Concentration (nM) | Volume (μL) | Final Concentration (nM) | |
| qScript XLT One-Step RT-qPCR ToughMix (2×) | 12.5 | 1× | / | / |
| Fluorescein sodium salt (1 μM) | 2.5 | 100 | / | / |
| One-Step RT-PCR Buffer (2×) | / | / | 12.5 | 1× |
| Ex Taq HS (5 μM) | / | / | 0.5 | 100 |
| Primer Script RT Enzyme Mix (5 μM) | / | / | 0.5 | 100 |
| C-PRRSV-F (25 μM) | 0.4 | 400 | 0.3 | 300 |
| C-PRRSV-R (25 μM) | 0.4 | 400 | 0.3 | 300 |
| C-PRRSV-P (25 μM) | 0.3 | 300 | 0.2 | 200 |
| HP-PRRSV-F (25 μM) | 1.0 | 1000 | 0.4 | 400 |
| HP-PRRSV-R (25 μM) | 1.0 | 1000 | 0.4 | 400 |
| HP-PRRSV-P (25 μM) | 0.5 | 500 | 0.5 | 500 |
| NL-PRRSV-F (25 μM) | 0.6 | 600 | 0.5 | 500 |
| NL-PRRSV-R (25 μM) | 0.6 | 600 | 0.5 | 500 |
| NL-PRRSV-P (25 μM) | 0.4 | 400 | 0.5 | 500 |
| Template (total nucleic acids) | 2.5 | / | 2.5 | / |
| RNase-free distilled water | Up to 25 | / | Up to 25 | / |
| Plasmid | Final Concentration (copies/µL) | Intra-Assay for Repeatability | Inter-Assay for Reproducibility | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Multiplex cdPCR (copies/µL) | Multiplex qRT-PCR(Ct) | Multiplex cdPCR (copies/µL) | Multiplex qRT-PCR (Ct) | ||||||||||
| SD | CV (%) | SD | CV (%) | SD | CV (%) | SD | CV (%) | ||||||
| p-C-PRRSV | 3.2 × 103 | 2334.33 | 37.45 | 1.68 | 24.171 | 0.05 | 0.19 | 2332.55 | 14.42 | 0.62 | 23.81 | 0.38 | 1.60 |
| 3.2 × 102 | 214.7 | 2.65 | 1.31 | 27.71 | 0.35 | 1.26 | 217.30 | 3.63 | 1.67 | 27.67 | 0.09 | 0.32 | |
| 3.2 × 101 | 19.10 | 0.25 | 1.70 | 30.89 | 0.18 | 0.58 | 18.90 | 0.38 | 2.01 | 30.67 | 0.52 | 1.70 | |
| p-HP-PRRSV | 3.2 × 103 | 2324 | 39.60 | 1.70 | 25.76 | 0.10 | 0.39 | 2310 | 57.20 | 2.48 | 25.64 | 0.10 | 0.39 |
| 3.2 × 102 | 196.5 | 3.90 | 1.98 | 29.34 | 0.21 | 0.72 | 201.4 | 5.30 | 2.63 | 29.54 | 0.19 | 0.64 | |
| 3.2 × 101 | 19.46 | 0.45 | 2.31 | 32.87 | 0.28 | 0.85 | 19.13 | 0.13 | 0.68 | 32.67 | 0.22 | 0.67 | |
| p-NL-PRRSV | 3.2 × 103 | 2328.3 | 37.07 | 1.59 | 24.28 | 0.26 | 1.07 | 2308.1 | 46.42 | 2.01 | 24.62 | 0.39 | 1.58 |
| 3.2 × 102 | 250.4 | 6.70 | 2.68 | 27.57 | 0.16 | 0.58 | 252.4 | 3.30 | 1.31 | 27.14 | 0.14 | 0.52 | |
| 3.2 × 101 | 23.2 | 0.55 | 2.37 | 30.92 | 0.12 | 0.39 | 22.86 | 0.57 | 2.49 | 30.74 | 0.24 | 0.78 | |
| Pathogen | Number | Multiplex qRT-PCR | Multiplex cdRT-PCR | Coincidence Rate (%) | Kappa | ||
|---|---|---|---|---|---|---|---|
| Positive | Positive Rate (%) | Positive | Positive Ratio (%) | ||||
| C-PRRSV | 320 | 6 | 1.88 | 7 | 2.19 | 99.69 | 0.92 |
| HP-PRRSV | 320 | 69 | 21.56 | 81 | 25.31 | 96.25 | 0.90 |
| NL-PRRSV | 320 | 31 | 9.69 | 37 | 11.56 | 98.13 | 0.90 |
| C-PRRSV plus HP-PRRSV | 320 | 2 | 0.63 | 2 | 0.63 | 100 | / |
| C-PRRSV plus NL-PRRSV | 320 | 1 | 0.31 | 1 | 0.31 | 100 | / |
| HP-PRRSV plus NL-PRRSV | 320 | 15 | 4.69 | 20 | 6.25 | 98.44 | / |
| C-PRRSV plus HP-PRRSV plus NL-PRRSV | 320 | 1 | 0.31 | 1 | 0.31 | 100 | / |
| Multiplex qRT-PCR | Multiplex cdRT-PCR | Coincidence Rate | Kappa Value | ||
|---|---|---|---|---|---|
| Positive | Negative | Total | |||
| Positive | 106 | 0 | 106 | 0.941 | 0.873 |
| Negative | 19 | 195 | 214 | ||
| Total | 125 | 195 | 320 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, F.; Chen, Y.; Shi, K.; Yin, Y.; Feng, S.; Si, H. Development of a Multiplex Crystal Digital RT-PCR for Differential Detection of Classical, Highly Pathogenic, and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus. Animals 2023, 13, 594. https://doi.org/10.3390/ani13040594
Long F, Chen Y, Shi K, Yin Y, Feng S, Si H. Development of a Multiplex Crystal Digital RT-PCR for Differential Detection of Classical, Highly Pathogenic, and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus. Animals. 2023; 13(4):594. https://doi.org/10.3390/ani13040594
Chicago/Turabian StyleLong, Feng, Yating Chen, Kaichuang Shi, Yanwen Yin, Shuping Feng, and Hongbin Si. 2023. "Development of a Multiplex Crystal Digital RT-PCR for Differential Detection of Classical, Highly Pathogenic, and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus" Animals 13, no. 4: 594. https://doi.org/10.3390/ani13040594
APA StyleLong, F., Chen, Y., Shi, K., Yin, Y., Feng, S., & Si, H. (2023). Development of a Multiplex Crystal Digital RT-PCR for Differential Detection of Classical, Highly Pathogenic, and NADC30-like Porcine Reproductive and Respiratory Syndrome Virus. Animals, 13(4), 594. https://doi.org/10.3390/ani13040594







