Molecular Detection of Porcine Parainfluenza Viruses 1 and 5 Using a Newly Developed Duplex Real-Time RT-PCR in South Korea
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Clinical Samples
2.2. Primers and Probes for dqRT-PCR
2.3. Reference Gene Construction for dqRT-PCR
2.4. Reference qRT-PCR Assays
2.5. Optimization of dqRT-PCR Conditions
2.6. Specificity and Sensitivity of dqRT-PCR
2.7. dqRT-PCR Precision
2.8. Clinical Evaluation of dqRT-PCR
2.9. Prevalence of PPIV1 and PPIV5 in Korean Pig Herds
3. Results
3.1. Interpretation of the dqRT-PCR Assay
3.2. Specificity of the dqRT-PCR Assay
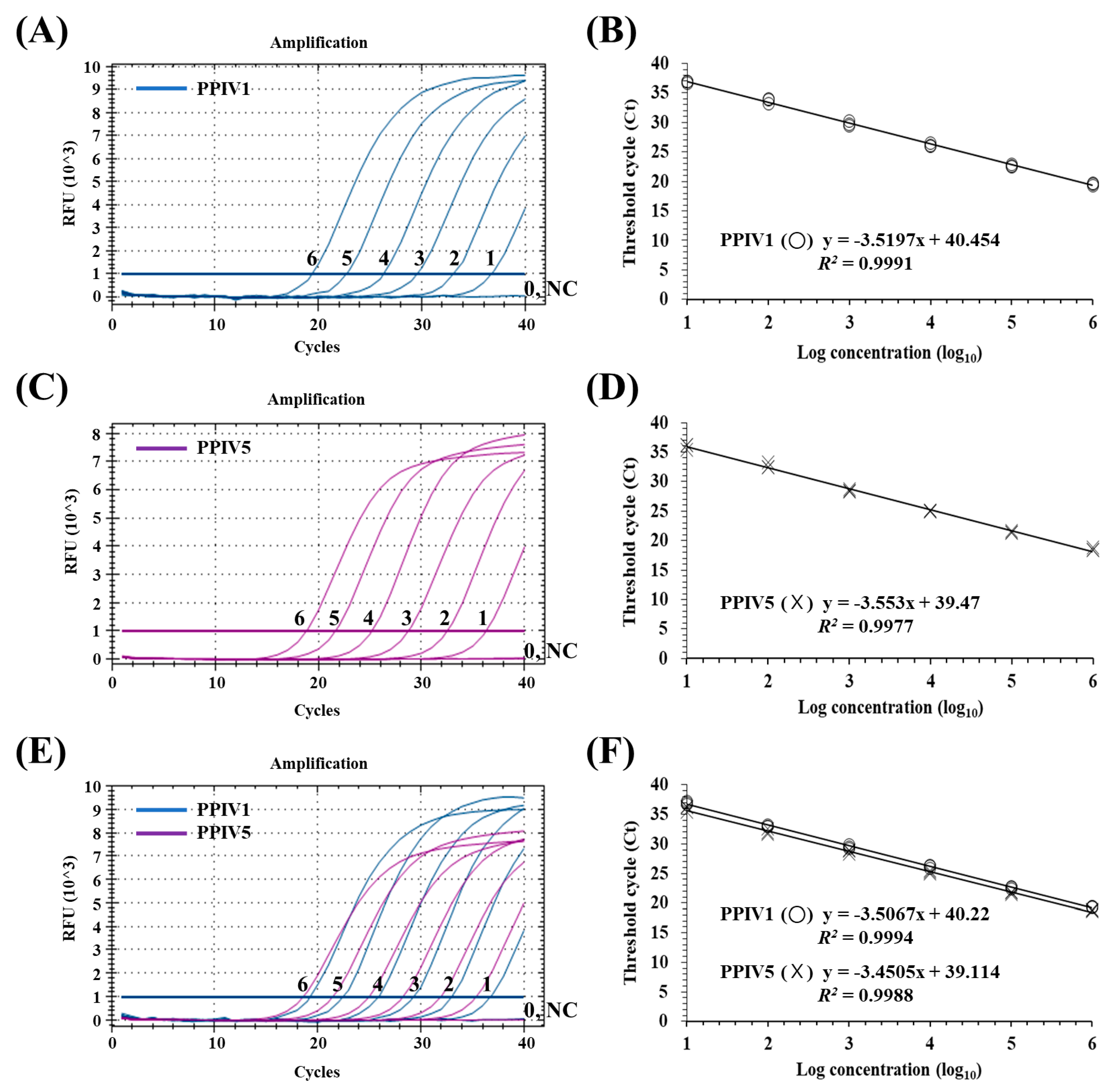
3.3. Sensitivity of the dqRT-PCR Assay
3.4. Precision of the dqRT-PCR Assay
3.5. Clinical Evaluation of the dqRT-PCR Assay
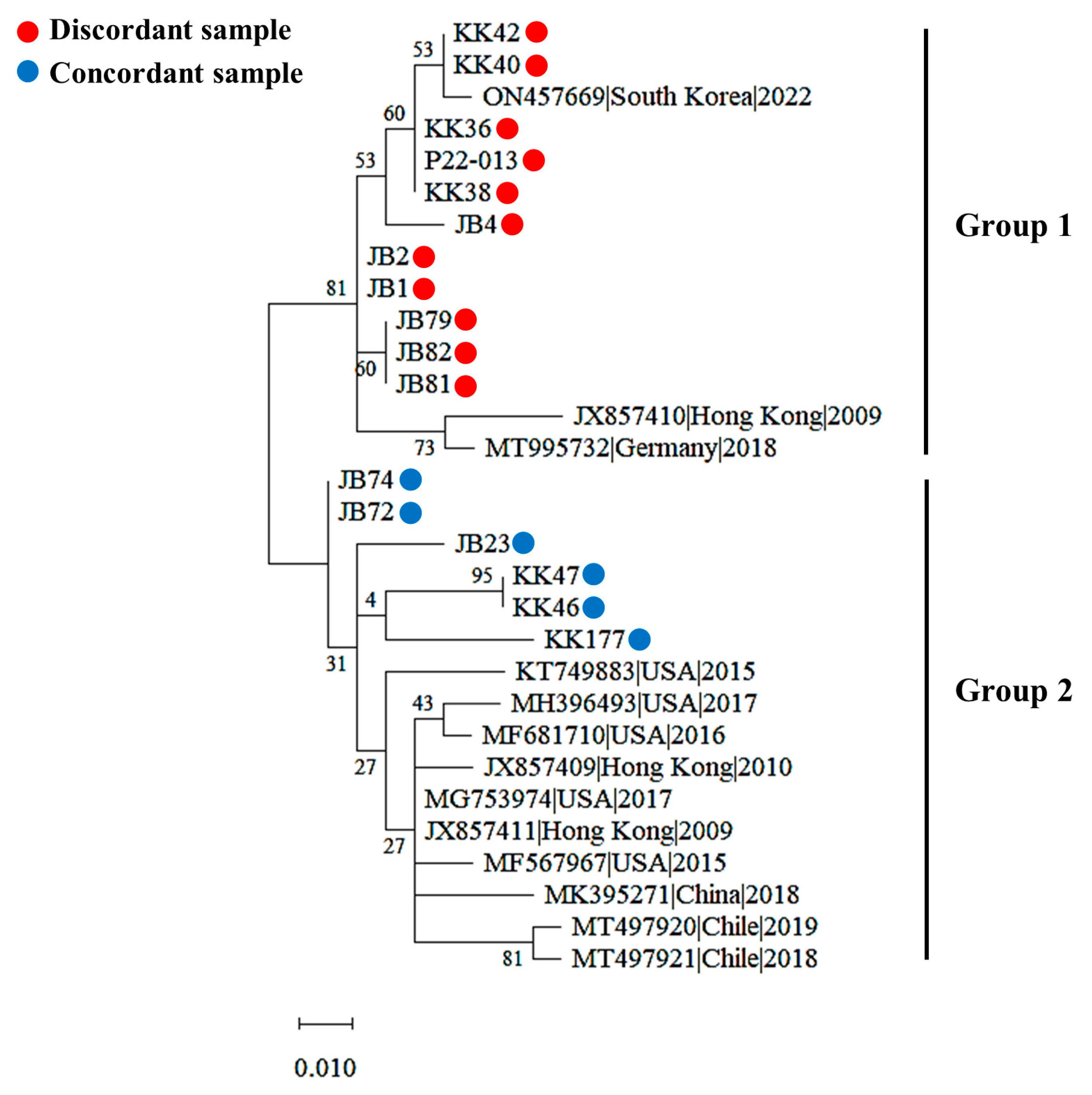
3.6. Genetic Diversity of PPIV1 Strains Based on the NP Gene
3.7. Prevalence of PPIV1 and PPIV5 in Korean Pig Herds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lau, S.K.P.; Woo, P.C.Y.; Wu, Y.; Wong, A.Y.P.; Wong, B.H.L.; Lau, C.C.Y.; Fan, R.Y.Y.; Cai, J.P.; Tsoi, H.W.; Chan, K.H.; et al. Identification and characterization of a novel paramyxovirus, porcine parainfluenza virus 1, from deceased pigs. J. Gen. Virol. 2013, 94, 2184–2190. [Google Scholar] [CrossRef]
- Agüero, B.; Mena, J.; Berrios, F.; Tapia, R.; Salinas, C.; Dutta, J.; van Bakel, H.; Mor, S.K.; Brito, B.; Medina, R.A.; et al. First report of porcine respirovirus 1 in South America. Vet. Microbiol. 2020, 246, 108726. [Google Scholar] [CrossRef] [PubMed]
- Dénes, L.; Cságola, A.; Schönhardt, K.; Halas, M.; Solymosi, N.; Balka, G. First report of porcine parainfluenza virus 1 (species Porcine respirovirus 1) in Europe. Transbound. Emerg. Dis. 2021, 68, 1731–1735. [Google Scholar] [CrossRef]
- Li, Y.; Sthal, C.; Bai, J.; Liu, X.; Anderson, G.; Fang, Y. Development of a real-time RT-qPCR assay for the detection of porcine respirovirus 1. J. Virol. Methods 2021, 289, 114040. [Google Scholar] [CrossRef] [PubMed]
- Palinski, R.M.; Chen, Z.; Henningson, J.N.; Lang, Y.; Rowland, R.R.R.; Fang, Y.; Prickett, J.; Gauger, P.C.; Hause, B.M. Widespread detection and characterization of porcine parainfluenza virus 1 in pigs in the USA. J. Gen. Virol. 2016, 97, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Welch, M.W.; Harmon, K.M.; Zhang, J.; Piñeyro, P.E.; Li, G.; Hause, B.M.; Gauger, P.C. Detection, isolation, and in vitro characterization of porcine parainfluenza virus type 1 isolated from respiratory diagnostic specimens in swine. Vet. Microbiol. 2019, 228, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, H.R.; Kim, J.M.; Lee, K.K.; Kim, W.I.; Lyoo, Y.S.; Kwon, O.D.; Park, C.K.; Park, S.C. First report of Porcine respirovirus 1 in South Korea. Transbound. Emerg. Dis. 2022, 69, 4041–4047. [Google Scholar] [CrossRef] [PubMed]
- Schuele, L.; Lizarazo-Forero, E.; Cassidy, H.; Strutzberg-Minder, K.; Boehmer, J.; Schuetze, S.; Loebert, S.; Lambrecht, C.; Harlizius, J.; Friedrich, A.W.; et al. First detection of porcine respirovirus 1 in Germany and the Netherlands. Transbound. Emerg. Dis. 2021, 68, 3120–3125. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, A.; Cybulski, P.; Denes, L.; Balka, G.; Stadejek, T. Detection of porcine respirovirus 1 (PRV1) in Poland: Incidence of co-Infections with influenza A virus (IAV) and porcine reproductive and respiratory syndrome Virus (PRRSV) in herds with a respiratory disease. Viruses 2022, 14, 148. [Google Scholar] [CrossRef]
- Rima, B.; Balkema-Buschmann, A.; Dundon, W.G.; Duprex, P.; Easton, A.; Fouchier, R.; Kurath, G.; Lamb, R.; Lee, B.; Rota, P.; et al. ICTV Virus Taxonomy Profile: Paramyxoviridae. J. Gen. Virol. 2019, 100, 1593–1594. [Google Scholar] [CrossRef]
- Heinen, E.; Herbst, W.; Schmeer, N. Isolation of a cytopathogenic virus from a case of porcine reproductive and respiratory syndrome (PRRS) and its characterization as parainfluenza virus type 2. Arch. Virol. 1998, 143, 2233–2239. [Google Scholar] [CrossRef] [PubMed]
- Chatziandreou, N.; Stock, N.; Young, D.; Andrejeva, J.; Hagmaier, K.; McGeoch, D.J.; Randall, R.E. Relationships and host range of human, canine, simian and porcine isolates of simian virus 5 (parainfluenza virus 5). J. Gen. Virol. 2004, 85, 3007–3016. [Google Scholar] [CrossRef]
- Jiang, N.; Wang, E.; Guo, D.; Wang, X.; Su, M.; Kong, F.; Yuan, D.; Zhai, J.; Sun, D. Isolation and molecular characterization of parainfluenza virus 5 in diarrhea-affected piglets in China. J. Vet. Med. Sci. 2018, 80, 590–593. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Lee, C. Complete genome sequence of a novel porcine parainfluenza virus 5 isolate in Korea. Arch. Virol. 2013, 158, 1765–1772. [Google Scholar] [CrossRef] [PubMed]
- Oem, J.K.; Kim, S.H.; Kim, Y.H.; Lee, M.H.; Lee, K.K. Molecular characteristics of canine parainfluenza viruses type 5 (CPIV-5) isolated in Korea. Can. J. Vet. Res. 2015, 79, 64–67. [Google Scholar] [PubMed]
- Xie, J.; Tong, P.; Zhang, A.; Zhang, L.; Song, X.; Kuang, L. Identification and characterization of the first equine parainfluenza Virus 5. Virol. Sin. 2020, 35, 245–247. [Google Scholar] [CrossRef]
- Zhai, J.Q.; Zhai, S.L.; Lin, T.; Liu, J.K.; Wang, H.X.; Li, B.; Zhang, H.; Zou, S.Z.; Zhou, X.; Wu, M.F.; et al. First complete genome sequence of parainfluenza virus 5 isolated from lesser panda. Arch. Virol. 2017, 162, 1413–1418. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.M.; Zhang, W.; Weird, G.M.; Zhang, H.; Pan, Y.; Zhang, L.; Xu, Y.; Li, C.; Chen, H.; Wang, Y. Characterization of parainfluenza virus 5 from diarrheic piglet highlights its zoonotic potential. Transbound. Emerg. Dis. 2022, 69, e1510–e1525. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.N.; Park, C.K.; Kim, S.H.; Lee, D.S.; Shin, J.H.; Lee, C. Characterization in vitro and in vivo of a novel porcine parainfluenza virus 5 isolate in Korea. Virus. Res. 2013, 178, 423–429. [Google Scholar] [CrossRef]
- Janke, B.H.; Paul, P.S.; Landgraf, J.G.; Halbur, P.G.; Huinker, C.D. Paramyxovirus infection in pigs with interstitial pneumonia and encephalitis in the United States. J. Vet. Diagn. Invest. 2001, 13, 428–433. [Google Scholar] [CrossRef] [Green Version]
- Qiao, D.; Janke, B.H.; Elankumaran, S. Molecular characterization of glycoprotein genes and phylogenetic analysis of two swine paramyxoviruses isolated from United States. Virus Genes 2009, 39, 53–65. [Google Scholar] [CrossRef]
- Qiao, D.; Janke, B.H.; Elankumaran, S. Complete genome sequence and pathogenicity of two swine parainfluenzavirus 3 isolates from pigs in the United States. J. Virol. 2010, 84, 686–694. [Google Scholar] [CrossRef]
- Park, J.Y.; Kim, S.H.; Lee, K.K.; Kim, Y.H.; Moon, B.Y.; So, B.; Park, C.K. Differential detection of porcine reproductive and respiratory syndrome virus genotypes by a fluorescence melting curve analysis using peptide nucleic acid probe-mediated one-step real-time RT-PCR. J. Virol. Methods 2019, 267, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Rima, B.K.; Gatherer, D.; Young, D.F.; Norsted, H.; Randall, R.E.; Davison, A.J. Stability of the parainfluenza virus 5 genome revealed by deep sequencing of strains isolated from different hosts and following passage in cell culture. J. Virol. 2014, 88, 3826–3836. [Google Scholar] [CrossRef]
- Navarro, E.; Serrano-Heras, G.; Castañoa, M.J.; Solera, J. Real-time PCR detection chemistry. Clin. Chim. Acta. 2015, 439, 231–250. [Google Scholar] [CrossRef]
- Dong, J.; Tsui, W.N.T.; Leng, X.; Fu, J.; Lohman, M.; Anderson, J.; Hamill, V.; Lu, N.; Porter, E.P.; Gray, M.; et al. Development of a three-panel multiplex real-time PCR assay for simultaneous detection of nine canine respiratory pathogens. J. Microbiol. Methods 2022, 199, 106528. [Google Scholar] [CrossRef] [PubMed]
- Hierweger, M.M.; Werder, S.; Seuberlich, T. Parainfluenza virus 5 infection in neurological disease and encephalitis of cattle. Int. J. Mol. Sci. 2020, 21, 498. [Google Scholar] [CrossRef]
- Broeders, S.; Huber, I.; Grohmann, L.; Berden, G.; Taverniers, I.; Mazzara, M.; Roosens, N.; Morisset, D. Guidelines for validation of qualitative real-time PCR methods. Trends. Food. Sci. Technol. 2014, 37, 115–126. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kwiecien, R.; Kopp-Schneider, A.; Blettner, M. Concordance analysis: Part 16 of a series on evaluation of scientific publications. Dtsch. Arztebl. Int. 2011, 108, 515–521. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mo.l Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tajadini, M.; Panjehpour, M.; Javanmard, S.H. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv. Biomed. Res. 2014, 3, 85. [Google Scholar] [CrossRef]
- Lamb, R.A.; Kolakofsky, D. Paramyxoviridae: The viruses and their replication. In Fields Virology, 4th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001; Volume 1, pp. 1305–1340. [Google Scholar]
- Hu, A.; Colella, M.; Zhao, P.; Li, F.; Tam, J.S.; Rappaport, R.; Cheng, S.M. 2005. Development of a real-time RT-PCR assay for detection and quantitation of parainfluenza virus 3. J. Virol. Methods 2005, 130, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Benitez, J.F.; García-Contreras, A.; Reyes-Leyva, J.; Hernández, J.; Sánchez-Betancourt, J.I.; Ramírez-Mendoza, H. Efficacy of quantitative RT-PCR for detection of the nucleoprotein gene from different porcine rubulavirus strains. Arch. Virol. 2013, 158, 1849–1856. [Google Scholar] [CrossRef]
- Klungthong, C.; Chinnawirotpisan, P.; Hussem, K.; Phonpakobsin, T.; Manasatienkij, W.; Ajariyakhajorn, C.; Rungrojcharoenkit, K.; Gibbons, R.V.; Jarman, R.G. The impact of primer and probe-template mismatches on the sensitivity of pandemic influenza A/H1N1/2009 virus detection by real-time RT-PCR. J. Clin. Virol. 2010, 48, 91–95. [Google Scholar] [CrossRef]
- Lengerova, M.; Racil, Z.; Volfova, P.; Lochmanova, J.; Berkovcova, J.; Dvorakova, D.; Vorlicek, J.; Mayer, J. Real-time PCR diagnostics failure caused by nucleotide variability within exon 4 of the human cytomegalovirus major immediate-early gene. J. Clin. Microbiol. 2007, 45, 1042–1044. [Google Scholar] [CrossRef]
- Süß, B.; Flekna, G.; Wagner, M.; Hein, I. Studying the effect of single mismatches in primer and probe binding regions on amplification curves and quantification in real-time PCR. J. Microbiol. Methods 2009, 76, 316–319. [Google Scholar]
- Yang, J.R.; Kuo, C.Y.; Huang, H.Y.; Wu, F.T.; Huang, Y.L.; Cheng, C.Y.; Su, Y.T.; Chang, F.Y.; Wu, H.S.; Liu, M.T. Newly Emerging Mutations in the Matrix Genes of the Human Influenza A(H1N1)pdm09 and A(H3N2) Viruses Reduce the Detection Sensitivity of Real-Time Reverse Transcription-PCR. J. Clin. Microbiol. 2014, 52, 76–82. [Google Scholar] [CrossRef]
- Welch, M.; Krueger, K.; Zhang, J.; Piñeyro, P.; Magtoto, R.; Wang, C.; Giménez-Lirola, L.; Strait, E.; Mogler, M.; Gauger, P. Detection of porcine parainfluenza virus type-1 antibody in swine serum using whole-virus ELISA, indirect fluorescence antibody and virus neutralizing assays. BMC. Vet. Res. 2022, 18, 110. [Google Scholar] [CrossRef]

| Pathogen a | Strain | Source b | Amplification of Target Gene | |
|---|---|---|---|---|
| PPIV1 (FAM) | PPIV5 (Cy5) | |||
| PPIV1 | KPPIV1-2201 | ADIC | + | - |
| PPIV5 | KPPIV5-2201 | ADIC | - | + |
| PCV2 | PCK0201 | ADIC | - | - |
| PCV3 | PCK3-1701 | ADIC | - | - |
| PRRSV-1 | Lelystad virus | APQA | - | - |
| PRRSV-2 | LMY | APQA | - | - |
| SIV | VDS1 | APQA | - | - |
| CSFV | LOM | APQA | - | - |
| PPV | NADL-2 | APQA | - | - |
| ST cell | - | ADIC | - | - |
| PK-15 cell | - | ADIC | - | - |
| Method | Primers /Probe | Sequence (5′–3′) a | Tm (°C ) | Genome Position b | Reference |
|---|---|---|---|---|---|
| dqRT-PCR for PPIV1 | P1-NF | ATAGAGGAGGTGGTGGTG | 59.8 | 178–195 | This study |
| P1-NR | GTGTTAARAACATGAGTGCTAT | 58.4 | 276–297 | ||
| P1-NP | FAM-TTATACCTGGACAAAAGAACACCGT-BHQ1 | 64.4 | 199–233 | ||
| dqRT-PCR for PPIV5 | P5-NF | CAACAGGGTGCAGTTGA | 59.4 | 1210–1226 | This study |
| P5-NR | GGTCAATTTRGCAAGTGTATT | 58.3 | 1282–1302 | ||
| P5-NP | Cy5-TCTCGGTCTAACTCAAGCCGAACGC-BHQ3 | 69.2 | 1245–1269 | ||
| qRT-PCR for PPIV1 | F | GCCAAAATGGCAGGGTTRTT | 63.4 | 114–133 | [8] |
| R | GCACCACCACCTCCTCTATT | 62.9 | 177–196 | ||
| P | FAM-TGCTCTCACTCCTTTTAGAACTAAATGTG-BHQ1 | 65 | 146–174 | ||
| qRT-PCR for PPIV5 | F | GATCATTCCGCTTAATCCCC | 60.3 | 425–444 | [26] |
| R | TTCTGCAAGTGCAGCATAGG | 62.7 | 482–501 | ||
| P | FAM-TCGTTCAGGTATGAGCCGTGGA-BHQ1 | 67 | 450–471 |
| Target Pathogen | Dilution (Copies/Reaction) | Intra-Assay | Inter-Assay | ||||
|---|---|---|---|---|---|---|---|
| Mean | SD | CV (%) | Mean | SD | CV (%) | ||
| PPIV1 | High (106) | 19.28 | 0.06 | 0.32 | 19.37 | 0.13 | 0.67 |
| Medium (104) | 25.98 | 0.1 | 0.4 | 26.11 | 0.17 | 0.66 | |
| Low (102) | 32.98 | 0.03 | 0.09 | 33.14 | 0.17 | 0.53 | |
| PPIV5 | High (106) | 18.60 | 0.1 | 0.55 | 18.72 | 0.15 | 0.8 |
| Medium (104) | 25.08 | 0.09 | 0.34 | 25.19 | 0.14 | 0.57 | |
| Low (102) | 31.68 | 0.09 | 0.29 | 32.04 | 0.46 | 1.44 | |
| Sample | No. of Tested | No. of Positive Agreements with the dqRT-PCR Assay (%) | No. of Positive Agreements with the Previous qRT-PCR Assays (%) | ||
|---|---|---|---|---|---|
| PPIV1 | PPIV5 | PPIV1 | PPIV5 | ||
| Lungs | 168 | 7 (4.2) | 2 (1.2) | 6 (3.6) | 2 (1.2) |
| Oral fluids | 120 | 60 (50.0) | 1 (0.8) a | 42 (35.0) | 1 (0.8) a |
| Sera | 153 | 0 | 0) | 0 | 0 |
| Total | 441 | 67 (15.2) | 3 (0.7) | 48 (10.9) | 3 (0.7) |
| Sample a | Sample Code | Sample Type | Assay Results (Ct Value) b | Sequencing | ||
|---|---|---|---|---|---|---|
| dqRT-PCR | qRT-PCR with Probe | qRT-PCR without Probe | ||||
| D1 | JB1 | Oral fluid | 32.45 | No Ct value | 29.13 | Yes |
| D2 | JB2 | Oral fluid | 31.32 | No Ct value | 28.51 | Yes |
| D3 | JB4 | Oral fluid | 32.09 | No Ct value | 30.54 | Yes |
| D4 | JB15 | Oral fluid | 35.09 | No Ct value | 32.65 | No |
| D5 | JB33 | Oral fluid | 34.68 | No Ct value | 31.59 | No |
| D6 | JB34 | Oral fluid | 36.59 | No Ct value | 33.92 | No |
| D7 | JB47 | Oral fluid | 35.17 | No Ct value | 32.41 | No |
| D8 | JB53 | Oral fluid | 33.09 | No Ct value | 30.14 | No |
| D9 | JB54 | Oral fluid | 33.73 | No Ct value | 30.03 | No |
| D10 | JB79 | Oral fluid | 30.67 | No Ct value | 29.24 | Yes |
| D11 | JB80 | Oral fluid | 35.39 | No Ct value | 32.98 | No |
| D12 | JB81 | Oral fluid | 26.52 | No Ct value | 25.4 | Yes |
| D13 | JB82 | Oral fluid | 31.18 | No Ct value | 28.98 | Yes |
| D14 | KK36 | Oral fluid | 27.22 | No Ct value | 26.14 | Yes |
| D15 | KK38 | Oral fluid | 28.04 | No Ct value | 27.71 | Yes |
| D16 | KK40 | Oral fluid | 28.26 | No Ct value | 30.45 | Yes |
| D17 | KK42 | Oral fluid | 27.18 | No Ct value | 25.02 | Yes |
| D18 | KK44 | Oral fluid | 35.09 | No Ct value | 33.09 | No |
| D19 | P22-013 | Lung | 27.41 | No Ct value | 26.55 | Yes |
| C1 | JB23 | Oral fluid | 27.87 | 31.81 | Not Tested | Yes |
| C2 | JB72 | Oral fluid | 28.01 | 28.94 | Not Tested | Yes |
| C3 | JB74 | Oral fluid | 27.02 | 28.13 | Not Tested | Yes |
| C4 | KK46 | Oral fluid | 28.95 | 31.31 | Not Tested | Yes |
| C5 | KK47 | Oral fluid | 28.27 | 29.95 | Not Tested | Yes |
| C6 | KK177 | Oral fluid | 30.85 | 35.76 | Not Tested | Yes |
| Pathogen | Farm-Level Prevalence | Pig-Level Prevalence | ||||
|---|---|---|---|---|---|---|
| No. of Tested | No. of Positive | % | No. of Tested | No. of Positive | % | |
| PPIV1 | 278 | 31 | 11.2 | 441 | 67 | 15.2 |
| PPIV5 | 278 | 3 | 1.1 | 441 | 3 | 0.7 |
| PPIV1 & 5 | 278 | 1 | 0.4 | 441 | 1 | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-M.; Kim, H.-R.; Jeon, G.-T.; Baek, J.-S.; Kwon, O.-D.; Park, C.-K. Molecular Detection of Porcine Parainfluenza Viruses 1 and 5 Using a Newly Developed Duplex Real-Time RT-PCR in South Korea. Animals 2023, 13, 598. https://doi.org/10.3390/ani13040598
Kim J-M, Kim H-R, Jeon G-T, Baek J-S, Kwon O-D, Park C-K. Molecular Detection of Porcine Parainfluenza Viruses 1 and 5 Using a Newly Developed Duplex Real-Time RT-PCR in South Korea. Animals. 2023; 13(4):598. https://doi.org/10.3390/ani13040598
Chicago/Turabian StyleKim, Jong-Min, Hye-Ryung Kim, Gyu-Tae Jeon, Ji-Su Baek, Oh-Deog Kwon, and Choi-Kyu Park. 2023. "Molecular Detection of Porcine Parainfluenza Viruses 1 and 5 Using a Newly Developed Duplex Real-Time RT-PCR in South Korea" Animals 13, no. 4: 598. https://doi.org/10.3390/ani13040598
APA StyleKim, J.-M., Kim, H.-R., Jeon, G.-T., Baek, J.-S., Kwon, O.-D., & Park, C.-K. (2023). Molecular Detection of Porcine Parainfluenza Viruses 1 and 5 Using a Newly Developed Duplex Real-Time RT-PCR in South Korea. Animals, 13(4), 598. https://doi.org/10.3390/ani13040598





