Environmental DNA Assay for the Detection of the American Bullfrog (Lithobates catesbeianus) in the Early Stages of the Invasion in the Ebre Delta
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. The American Bullfrog
1.2. Environmental DNA (eDNA) for Monitoring the Process of the Invasion
2. Material and Methods
2.1. Study Area and Bullfrog Detection
2.2. eDNA Sampling and Extraction
2.3. eDNA Amplification
| Universal Amphibian | Primer Sequences | References |
|---|---|---|
| 16SA-L | CGCCTGTTTATCAAAAACAT | Vences et al. (2005) [41] |
| 16SB-H | CCGGTCTGAACTCAGATCACGT | Vences et al. (2005) [41] |
| American bullfrog | ||
| CytbF1 | TGCCAACGGAGCATCATTC | Ficetola et al. (2008a) [29] |
| CytbF2 | GTTAATAACGGCTGACTCCTA | This study |
| CytbR1 | ATAAAGGTAGGAGCCGTAGT | Ficetola et al. (2008a) [29] |
| CytbR2 | GATATTTGGCCCCATGGT | This study |
| Primer set | Fragment length | |
| CytbF1 + CytbR2 | 168 bp | |
| CytbF2 + CytbR1 | 111 bp | |
| CytbF1 + CytbR1 | 79 bp | |
| 16SA-L + 16SB-H | 594 bp |
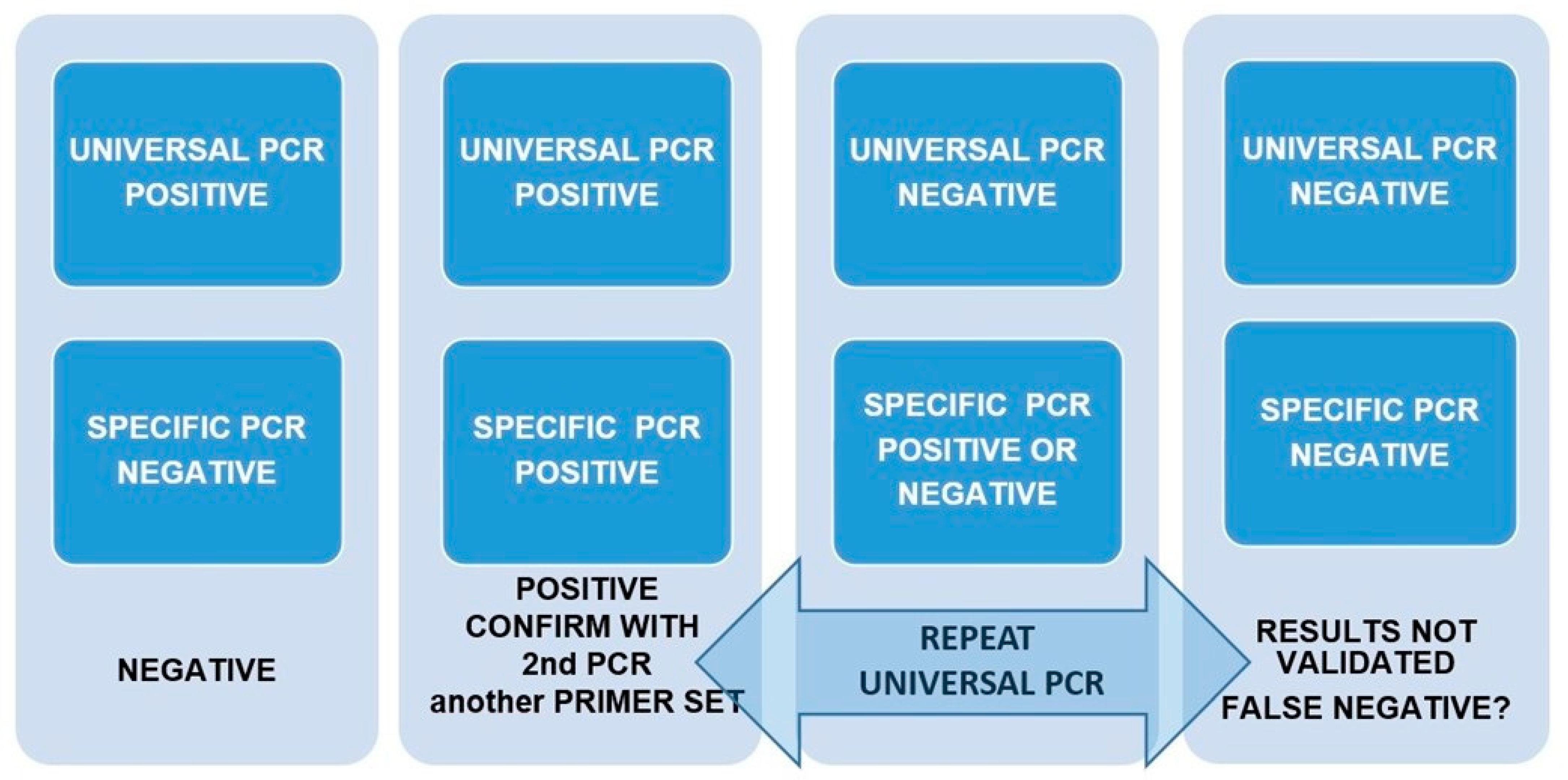
2.4. Design of a Pipeline to Validate Results in Front of the Possibility of False Positives and Negatives
3. Results
3.1. 2019 Survey
3.2. 2020 Surveys
3.2.1. June eDNA Survey
3.2.2. July eDNA Survey
4. Discussion
4.1. Challenges of eDNA to Track Invasive Species
4.1.1. Environmental DNA Capture
4.1.2. PCR and False Positive/Negative Results
4.1.3. Replicates and Threshold of Positivity
4.2. Early Invasion of the American Bullfrog in the Ebre Delta Revealed by eDNA
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehrenfeld, J.G. Ecosystem consequences of biological invasions. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 59–80. [Google Scholar] [CrossRef] [Green Version]
- Pyšek, P.; Richardson, D.M. Invasive species, environmental change and management, and health. Annu. Rev. Environ. Resour. 2010, 35, 25–55. [Google Scholar] [CrossRef] [Green Version]
- Garner, T.W.J.; Perkins, M.W.; Govindarajulu, P.; Seglie, D.; Walker, S.; Cunningham, A.; Fisher, M.C. The emerging amphibian pathogen Batrachochytrium dendrobatidis globally infects introduced populations of the North American bullfrog, Rana catesbeiana. Biol. Lett. 2006, 2, 455–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SIARE. Base de Datos de Anfibios y Reptiles de España. Available online: https://siare.herpetologica.es/index.php?/bdh/distribucion (accessed on 6 June 2022).
- Darwall, W.; Carrizo, S.; Numa, C.; Barrios, V.; Freyhof, J.; Smith, K.; Freshwater Key Biodiversity Areas in the Mediterranean Basin Hotspot. Technical report IUCN. 2014. Available online: https://portals.iucn.org/library/sites/library/files/documents/SSC-OP-052.pdf (accessed on 1 June 2022).
- Fatorić, S.L.; Chelleri, L. Vulnerability to the effects of climate change and adaptation: The case of the Spanish Ebre Delta. Ocean Coast. Manage. 2012, 60, 1–10. [Google Scholar] [CrossRef]
- Peña-Gallardo, M.; Vicente-Serrano, S.M.; Domínguez-Castro, F.; Beguería, S. The impact of drought on the productivity of two rainfed crops in Spain. Nat. Hazards Earth Syst. Sci. 2019, 19, 1215–1234. [Google Scholar] [CrossRef] [Green Version]
- Global Invasive Species Database. 2022. Available online: http://www.iucngisd.org/gisd/100_worst.phpon21-12-2022 (accessed on 1 June 2022).
- Capinha, C.; Seebens, H.; Cassey, P.; García-Díaz, P.; Lenzner, B.; Mang, T.; Moser, D.; Pyšek, P.; Rödder, D.; Scalera, R.; et al. Diversity, biogeography and the global flows of alien amphibians and reptiles. Divers. Distrib. 2017, 23, 1313–1322. [Google Scholar] [CrossRef] [Green Version]
- Jancowski, K.; Orchard, S. Stomach contents from invasive American bullfrogs Rana catesbeiana (= Lithobates catesbeianus) on southern Vancouver Island, British Columbia, Canada. NeoBiota 2013, 16, 17–37. [Google Scholar] [CrossRef] [Green Version]
- Yap, T.A.; Nguyen, N.T.; Serr, M.; Shepack, A.; Vredenburg, V.T. Batrachochytrium salamandrivorans and the Risk of a Second Amphibian Pandemic. Ecohealth 2017, 14, 851–864. [Google Scholar] [CrossRef]
- Longcore, J.E.; Pessier, A.P.; Nichols, D.K. Batrachochytrium Dendrobatidis Gen. et Sp. Nov., a Chytrid Pathogenic to Amphibians. Mycologia 1999, 91, 219–227. [Google Scholar] [CrossRef]
- Urbina, J.; Bredeweg, E.M.; Garcia, T.S.; Blaustein, A.R. Host–pathogen dynamics among the invasive American bullfrog (Lithobates catesbeianus) and chytrid fungus (Batrachochytrium dendrobatidis). Hydrobiologia 2018, 817, 267–277. [Google Scholar] [CrossRef]
- Bosch, J.; Martínez-Solano, I.; García-París, M. Evidence of a chytrid fungus infection involved in the decline of the common midwife toad (Alytes obstetricans) in protected areas of central Spain. Biol. Conserv. 2001, 97, 331–337. [Google Scholar] [CrossRef]
- Pounds, J.A.; Bustamante, M.R.; Coloma, L.A.; Consuegra, J.A.; Fogden, M.P.L.; Foster, P.N.; La Marca, E.; Masters, K.L.; Merino-Viteri, A.; Puschendorf, R.; et al. Widespread amphibian extinctions from epidemic disease driven by global warming. Nature 2006, 439, 161–167. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Coïc, C.; Detaint, M.; Berroneau, M.; Lorvelec, O.; Miaud, C. Pattern of distribution of the American bullfrog Rana catesbeiana in Europe. Biol. Invasions 2007, 9, 767–772. [Google Scholar] [CrossRef]
- Kraus, F. Impacts of Alien Reptiles and Amphibians. In Alien Reptiles and Amphibians; Kraus, F., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 57–93. [Google Scholar] [CrossRef]
- Urioste, J.; Bethencourt, M.J. Rana toro y Sapo Marino: La amenaza que viene. Rev. Medio Ambiente Canar. 2001, 21, 26–30. [Google Scholar]
- Pleguezuelos, J.M. Las Especies introducidas de Anfibios y Reptiles. In Atlas y Libro Rojo los Anfibios y Reptil España; Pleguezuelos, J.M., Márquez, R., Lizana, M., Eds.; Ministerio de Medio Ambiente, Gobierno de España: Madrid, Spain, 2002; pp. 501–532. [Google Scholar]
- Cabana, M.; Fernández, D. Nueva vía de entrada de rana toro (Lithobates catesbeianus) en la Península Ibérica. Boletín Asoc. Herpetológica Española 2010, 21, 101–104. [Google Scholar]
- Franch, N. Cita de granota toro (Lithobates catesbeianus) al Delta de l’Ebre. Herpetofull Soc. Catalana Herpetol. 2012, 8, 14–15. [Google Scholar]
- Blackburn, T.M.; Pyšek, P.; Bacher, S.; Carlton, J.T.; Duncan, R.P.; Jarošík, V.; Wilson, J.R.U.; Richardson, D.M. A proposed unified framework for biological invasions. Trends Ecol. Evol. 2011, 26, 333–339. [Google Scholar] [CrossRef] [Green Version]
- Palau, A.; Cía, I. Métodos de Control y Eradicación del Mejillón Cebra (Dreissena polymorpha). Dirección y Coordinación: A Palau, E Rosico; Dirección de Medio Ambiente y Desarrollo Sostenible, Endesa: Madrid, Spain, 2006. [Google Scholar]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Anderson, L.W.J. California’s reaction to Caulerpa taxifolia: A model for invasive species rapid response. Biol. Invasions 2005, 7, 1003–1016. [Google Scholar] [CrossRef]
- Wimbush, J.; Frischer, M.E.; Zarzynski, J.W.; Nierzwicki-Bauer, S.A. Eradication of colonizing populations of zebra mussels (Dreissena polymorpha) by early detection and SCUBA removal: Lake George, NY. Aquat. Conserv.-Mar. Freshw. Ecosyst. 2009, 19, 703–713. [Google Scholar] [CrossRef]
- Magnuson, J.J.; Benson, B.J.; Mclain, A.S. Insights on Species Richness and Turnover from Long-Term Ecological Research: Fishes in North Temperate Lakes. Am. Zool. 1994, 34, 437–451. [Google Scholar] [CrossRef] [Green Version]
- Schill, W.B.; Galbraith, H.S. Detecting the undetectable: Characterization, optimization, and validation of an eDNA detection assay for the federally endangered dwarf wedgemussel, Alasmidonta heterodon (Bivalvia: Unionoida). Aquat. Conserv.-Mar. Freshw. Ecosyst 2019, 29, 603–611. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4, 423–425. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.P.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Møller, P.R.; Rasmussen, M.; Willerslev, E. Detection of a Diverse Marine Fish Fauna Using Environmental DNA from Seawater Samples. PLoS One 2012, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, M.W.; Overballe-Petersen, S.; Ermini, L.; Sarkissian, C.D.; Haile, J.; Hellstrom, M.; Spens, J.; Thomsen, P.F.; Bohmann, K.; Cappellini, E.; et al. Ancient and modern environmental DNA. Philos. Trans. R. Soc. B-Biol. Sci. 2015, 370, 20130383. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, C.S.; Strickler, K.M.; Fremier, A.K. Degradation and Dispersion Limit Environmental DNA Detection of Rare Amphibians in Wetlands: Increasing Efficacy of Sampling Designs. Sci. Total Environ. 2018, 633, 695–703. [Google Scholar] [CrossRef]
- Torresdal, J.D.; Farrell, A.D.; Goldberg, C.S. Environmental DNA Detection of the Golden Tree Frog (Phytotriades auratus) in Bromeliads. PLoS ONE 2017, 12, e0168787. [Google Scholar] [CrossRef] [Green Version]
- Dejean, T.; Valentini, A.; Miquel, C.; Taberlet, P.; Bellemain, E.; Miaud, C. Improved detection of an alien invasive species through environmental DNA barcoding: The example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 2012, 49, 953–959. [Google Scholar] [CrossRef]
- Kamoroff, C.; Daniele, N.; Grasso, R.L.; Rising, R.; Espinoza, T.; Goldberg, C.S. Effective removal of the American bullfrog (Lithobates catesbeianus) on a landscape level: Long term monitoring and removal efforts in Yosemite Valley, Yosemite National Park. Biol. Invasions 2020, 22, 617–626. [Google Scholar] [CrossRef] [Green Version]
- Majaneva, M.; Diserud, O.H.; Eagle, S.H.C.; Boström, E.; Hajibabaei, M.; Ekrem, T. Environmental DNA filtration techniques affect recovered biodiversity. Sci. Rep. 2018, 8, 4682. [Google Scholar] [CrossRef]
- Llorente-Cabrera, G.; Montori-Faura, A.; Pujol-Buxó, E. El sapillo pintojo mediterráneo (Discoglossus pictus) en la península ibérica. Bol. Asoc. Herpetol. Esp. 2015, 26, 12–17. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [Green Version]
- Roig-Cort, J.; Guillem-Giner, P.; Gómez-García, D. Noves aportacions a la distribució dels amfibis i els rèptils del sud de Catalunya. Butlletí Soc. Catatalana D’herpetologia 2012, 20, 26–36. [Google Scholar]
- Vences, M.; Thomas, M.; Van Der Meijden, A.; Chiari, Y.; Vieites, D.R. Comparative performance of the 16S rRNA gene in DNA barcoding of amphibians. Front. Zool. 2005, 2, 5. [Google Scholar] [CrossRef] [Green Version]
- Smart, A.S.; Tingley, R.; Weeks, A.R.; Van Rooyen, A.R.; McCarthy, M.A. Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol. Appl. 2015, 25, 1944–1952. [Google Scholar] [CrossRef]
- Baudry, T.; Mauvisseau, Q.; Goût, J.P.; Arqué, A.; Delaunay, C.; Smith-Ravin, J.; Sweet, M.; Grandjean, F. Mapping a super-invader in a biodiversity hotspot, an eDNA-based success story. Ecol. Indic. 2021, 126, 107637. [Google Scholar] [CrossRef]
- Everts, T.; Halfmaerten, D.; Neyrinck, S.; De Regge, N.; Jacquemyn, H.; Bryset, R. Accurate detection and quantification of seasonal abundance of American bullfrog (Lithobates catesbeianus) using ddPCR eDNA assays. Sci. Rep. 2021, 11, 11282. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef]
- Takahashi, S.; Sakata, M.K.; Minamoto, T.; Masuda, R. Comparing the efficiency of open and enclosed filtration systems in environmental DNA quantification for fish and jellyfish. PLoS ONE 2020, 15, e0231718. [Google Scholar] [CrossRef] [Green Version]
- Hunter, M.E.; Ferrante, J.A.; Meigs-Friend, G.; Ulmer, A. Improving eDNA yield and inhibitor reduction through increased water volumes and multi-filter isolation techniques. Sci. Rep. 2019, 9, 5259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeunen, G.J.; Knapp, M.; Spencer, H.G.; Taylor, H.R.; Lamare, M.D.; Stat, M.; Bunce, M.; Gemmell, N.J. Species-level biodiversity assessment using marine environmental DNA metabarcoding requires protocol optimization and standardization. Ecol. Evol. 2019, 9, 1323–1335. [Google Scholar] [CrossRef] [Green Version]
- Deagle, B.E.; Eveson, J.P.; Jarman, S.N. Quantification of damage in DNA recovered from highly degraded samples—A case study on DNA in faeces. Front. Zool. 2006, 3, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahara, T.; Minamoto, T.; Doi, H. Using Environmental DNA to Estimate the Distribution of an Invasive Fish Species in Ponds. PLoS ONE 2013, 8, e56584. [Google Scholar] [CrossRef] [Green Version]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Jane, S.F.; Lowe, W.H.; Whiteley, A.R.; Schwartz, M.K. Robust Detection of Rare Species Using Environmental DNA: The Importance of Primer Specificity. PLoS ONE 2013, 8, e59520. [Google Scholar] [CrossRef] [Green Version]
- McKee, A.M.; Spear, S.F.; Pierson, T.W. The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biol. Conserv. 2015, 183, 70–76. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef] [Green Version]
- Ficetola, G.F.; Pansu, J.; Bonin, A.; Coissac, E.; Giguet-Covex, C.; De Barba, M.; Gielly, L.; Lopes, C.M.; Boyer, F.; Raye, F.P.G.; et al. Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 2015, 15, 543–556. [Google Scholar] [CrossRef]
- Taberlet, P.; Griffin, S.; Goossens, B.; Questiau, S.; Manceau, V.; Escaravage, N.; Waits, L.P.; Bouvet, J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996, 24, 3189–3194. [Google Scholar] [CrossRef] [Green Version]
- Sepulveda, A.J.; Hutchins, P.R.; Forstchen, M.; Mckeefry, M.N.; Swigris, A.M. The Elephant in the Lab (and Field): Contamination in Aquatic Environmental DNA Studies. Front. Ecol. Evol. 2020, 8, 1–12. [Google Scholar] [CrossRef]
- Kamath, P.L.; Sepulveda, A.J.; Layhee, M. Genetic reconstruction of a bullfrog invasion to elucidate vectors of introduction and secondary spread. Ecol. Evol. 2016, 6, 5221–5233. [Google Scholar] [CrossRef] [Green Version]
- Farré, M.; Lombarte, A.; Tuset, V.M.; Abelló, P. Shape matters: Relevance of carapace for brachyuran crab invaders. Biol. Invasions 2021, 23, 461–475. [Google Scholar] [CrossRef]
- Blackburn, T.M.; Lockwood, J.L.; Cassey, P. The influence of numbers on invasion success. Mol. Ecol. 2015, 24, 1942–1953. [Google Scholar] [CrossRef] [Green Version]
- Ficetola, G.F.; Bonin, A.; Miaud, C. Population genetics reveals origin and number of founders in a biological invasion. Mol. Ecol. 2008, 17, 773–782. [Google Scholar] [CrossRef]
- Suarez, A.V.; Tsutsui, N.D. The evolutionary consequences of biological invasions. Mol. Ecol. 2008, 17, 351–360. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Thuiller, W.; Miaud, C. Prediction and validation of the potential global distribution of a problematic alien invasive species—The American bullfrog. Divers. Distrib. 2007, 13, 476–485. [Google Scholar] [CrossRef]
- Gilioli, G.; Schrader, G.; Carlsson, N.; vanDonk, E.; van Leeuwen, C.H.A.; Martín, P.R.; Pasquali, S.; Vilà, M.; Vos, S. Environmental risk assessment for invasive alien species: A case study of apple snails affecting ecosystem services in Europe. Environ. Impact Assess. Rev. 2017, 65, 1–11. [Google Scholar] [CrossRef]
- Zografos, C. Flows of sediment, flows of insecurity: Climate change adaptation and the social contract in the Ebro Delta, Catalonia. Geoforum 2017, 80, 49–60. [Google Scholar] [CrossRef]
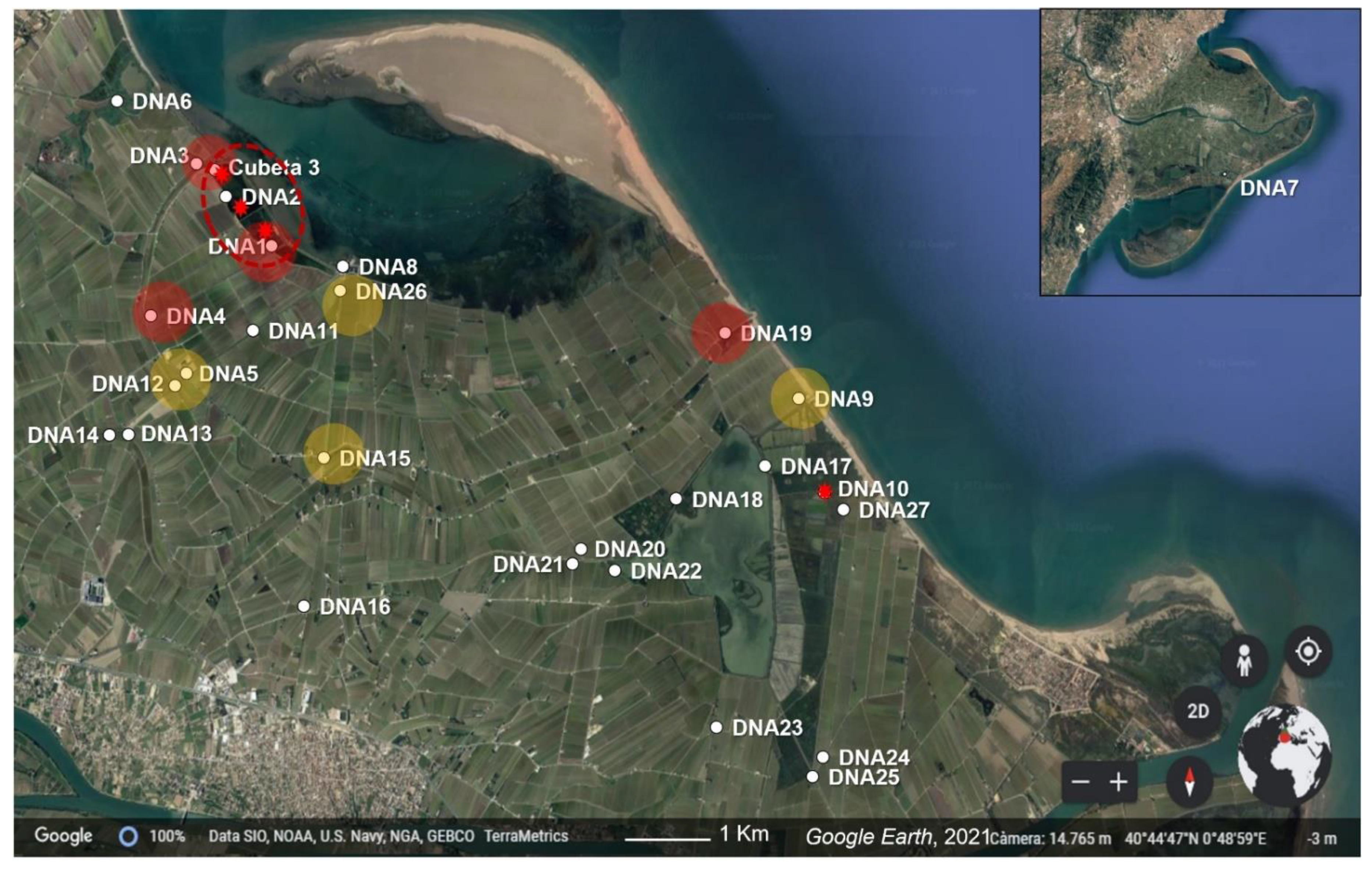
| 2018 | 2019 | 2020 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sampling Effort | Number of Captures | Period | Sampling Effort | Number of Captures | Period | Sampling Effort | Number of Captures | Period | |
| Terrestrial transects | 73 | 76 | August–November | 257 | 0 | January–November | 195 | 2 | April–October |
| Acoustic surveys | 15 | 3 | June–July | 80 | 0 | April–November | 39 | 1 | June–August |
| Tadpole coop traps | 235 | 1057 | June–October | 792 | 0 | May–September | 704 | 0 | May–October |
| Adult traps | - | 90 | 0 | July–September | 150 | 0 | June–October | ||
| eDNA | - | 8 | 0 | July | 32 | 10 | June–July | ||
| Total Positives/Total Number of Replicates | First Round of PCR F1 + R2 | Second Round of PCR F1 + R2 | Alternative PCR F2 + R1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| June survey | F1 + R2 | 1:10 (3) | 1:100 (3) | 1:10 (2) | 1:100 (3) | 1:1000 (3) | |||||
| DNA3 | NEG | NEG | NEG | ||||||||
| DNA4* | NEG | NEG | NEG | NEG | NEG | NEG | |||||
| DNA5* | 3/14 | NEG | NEG | NEG | 2/3 | 1/3 | |||||
| DNA6 | NEG | NEG | NEG | ||||||||
| DNA7 | NEG | NEG | NEG | ||||||||
| DNA8 | NEG | NEG | NEG | NEG | NEG | NEG | |||||
| DNA9* | 5/14 | NEG | 1/3 | 1/2 | 2/3 | 1/3 | |||||
| DNA10 | NEG | NEG | NEG | NEG | NEG | NEG | |||||
| July survey | F1 + R2 | F2 + R1 | 1:10 (3) | 1:100 (3) | 1:1000 (2) | 1:10 (2) | 1:100 (3) | 1:1000 (3) | 1:10 (2) | 1:100 (3) | 1:1000 (3) |
| DNA1 | 1/16 | 1/8 | NEG | NEG | 1/2 | NEG | NEG | NEG | NEG | NEG | 1/3 |
| DNA3 | 1/16 | 2/8 | NEG | 1/3 | NEG | NEG | NEG | NEG | NEG | NEG | 2/3 |
| DNA4 | 4/16 | 2/8 | NEG | NEG | 1/2 | NEG | 1/3 | 2/3 | NEG | NEG | 2/3 |
| DNA5 | 5/16 | NEG | NEG | NEG | NEG | NEG | 3/3 | 2/3 | NEG | NEG | NEG |
| DNA6 | NEG | NEG | NEG | ||||||||
| DNA7 | NEG | NEG | NEG | ||||||||
| DNA8 | NEG | NEG | NEG | ||||||||
| DNA9 | 1/16 | NEG | NEG | NEG | NEG | NEG | 1/3 | NEG | NEG | NEG | NEG |
| DNA10 | NEG | NEG | NEG | ||||||||
| DNA11 | NEG | NEG | NEG | NEG | |||||||
| DNA12 | 2/16 | NEG | NEG | NEG | 1/2 | NEG | NEG | 1/3 | NEG | NEG | NEG |
| DNA13 | NEG | NEG | NEG | NEG | |||||||
| DNA14 | NEG | NEG | NEG | NEG | |||||||
| DNA15 | 2/16 | NEG | NEG | 1/3 | 1/2 | NEG | NEG | NEG | NEG | NEG | NEG |
| DNA16 | NEG | NEG | NEG | NEG | |||||||
| DNA17 | NEG | NEG | NEG | NEG | |||||||
| DNA18 | NEG | NEG | NEG | NEG | |||||||
| DNA19 | 2/16 | 1/8 | NEG | NEG | 1/2 | NEG | 1/3 | NEG | NEG | 1/3 | NEG |
| DNA20 | NEG | NEG | NEG | NEG | |||||||
| DNA21 | NEG | NEG | NEG | NEG | |||||||
| DNA22* | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG |
| DNA23* | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | NEG | |
| DNA24 | NEG | NEG | NEG | NEG | |||||||
| DNA25 | NEG | NEG | NEG | NEG | |||||||
| DNA26* | 2/16 | NEG | NEG | 1/3 | 1/2 | NEG | NEG | NEG | NEG | NEG | NEG |
| DNA27 | NEG | NEG | NEG | NEG | |||||||
| cubeta3 | 6/16 | 5/8 | 3/3 | 2/3 | NEG | NEG | 1/3 | NEG | NEG | 3/3 | 2/3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanz, N.; Franch, N.; Araguas, R.-M.; Viñas, J.; Vidal, O. Environmental DNA Assay for the Detection of the American Bullfrog (Lithobates catesbeianus) in the Early Stages of the Invasion in the Ebre Delta. Animals 2023, 13, 683. https://doi.org/10.3390/ani13040683
Sanz N, Franch N, Araguas R-M, Viñas J, Vidal O. Environmental DNA Assay for the Detection of the American Bullfrog (Lithobates catesbeianus) in the Early Stages of the Invasion in the Ebre Delta. Animals. 2023; 13(4):683. https://doi.org/10.3390/ani13040683
Chicago/Turabian StyleSanz, Nuria, Nati Franch, Rosa-Maria Araguas, Jordi Viñas, and Oriol Vidal. 2023. "Environmental DNA Assay for the Detection of the American Bullfrog (Lithobates catesbeianus) in the Early Stages of the Invasion in the Ebre Delta" Animals 13, no. 4: 683. https://doi.org/10.3390/ani13040683






