Effect of Spirulina Dietary Supplementation in Modifying the Rumen Microbiota of Ewes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Diets and Experimental Design
2.2. Sample Collection
2.3. DNA Extraction
2.4. Primer Design and Relative Abundance of Selected Rumen Microorganisms
2.5. Metagenomic NGS Analysis
2.6. Statistical Analysis
3. Results
3.1. 16S Amplicon Sequencing
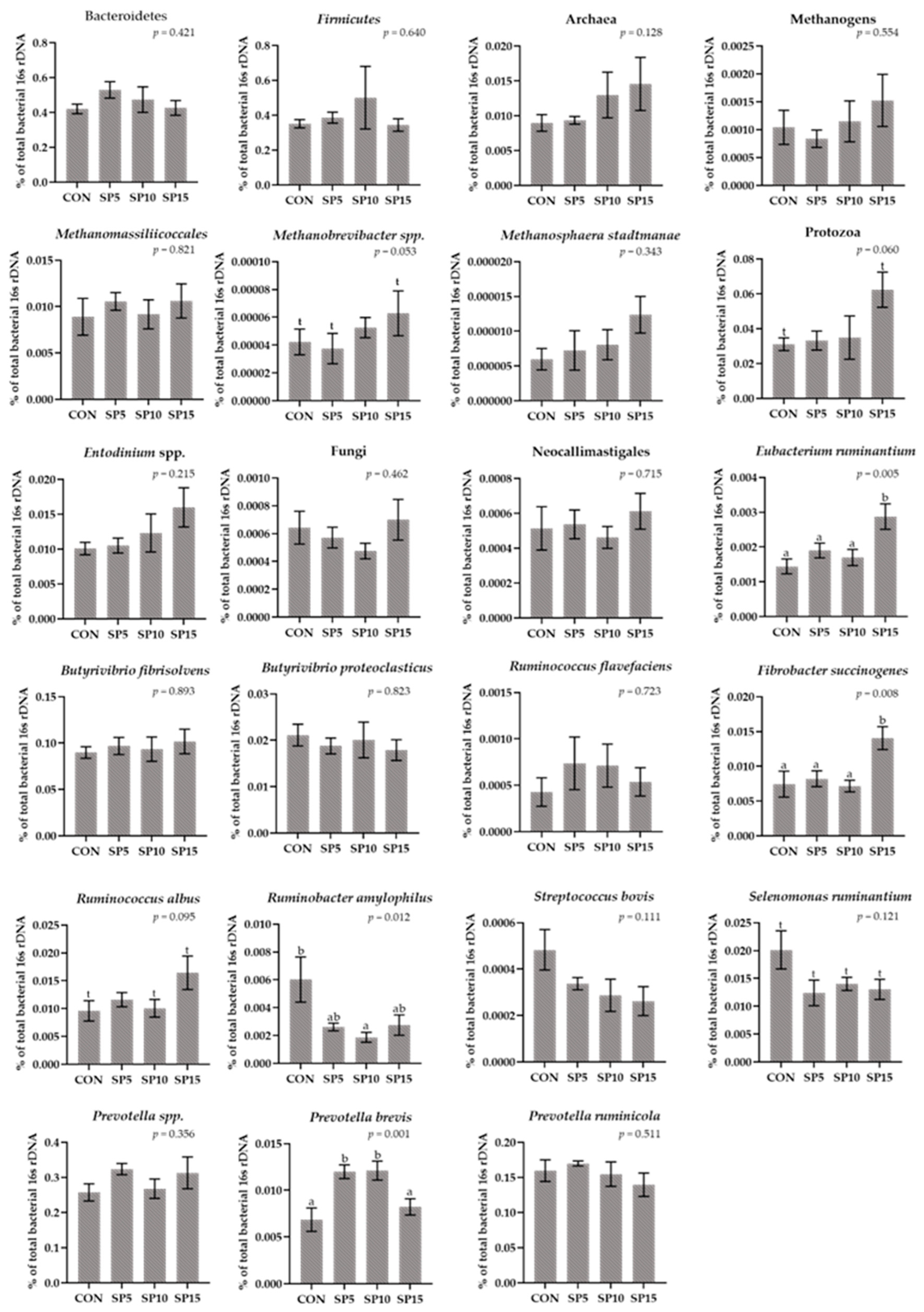
3.2. Relative Abundance of Selected Microorganisms in the Rumen Fluid Samples Using a RT-qPCR Platform
3.3. Relative Abundance of Selected Microorganisms in the Rumen Solid Particle Using RT-qPCR Platform
4. Discussion
The Spirulina Supplementation Demonstrated Important Findings in the Modification of the Populations of Rumen Microorganisms
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dineshbabu, G.; Goswami, G.; Kumar, R.; Sinha, A.; Das, D. Microalgae–nutritious, sustainable aqua- and animal feed source. J. Funct. Foods 2019, 62, 103545. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Abdullah, M.A.M.; Alexandros, M.; Chatzikonstantinou, M.; Skliros, D.; Sotirakoglou, K.; Flemetakis, E.; Labrou, N.E.; Zervas, G. The effect of dietary Chlorella pyrenoidosa inclusion on goats milk chemical composition, fatty acids profile and enzymes activities related to oxidation. Livest. Sci. 2017, 197, 106–111. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Abdullah, M.A.M.; Mavrommatis, A.; Chatzikonstantinou, M.; Skliros, D.; Sotirakoglou, K.; Flemetakis, E.; Labrou, N.E.; Zervas, G. The effect of dietary Chlorella vulgaris inclusion on goat’s milk chemical composition, fatty acids profile and enzymes activities related to oxidation. J. Anim. Physiol. Anim. Nutr. 2018, 102, 142–151. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Sotirakoglou, K.; Kamilaris, C.; Tsiplakou, E. Effects of Inclusion of Schizochytrium spp. and Forage-to-Concentrate Ratios on Goats’ Milk Quality and Oxidative Status. Foods 2021, 10, 1322. [Google Scholar] [CrossRef] [PubMed]
- Mavrommatis, A.; Chronopoulou, E.G.; Sotirakoglou, K.; Labrou, N.E.; Zervas, G.; Tsiplakou, E. The impact of the dietary supplementation level with schizochytrium sp, on the oxidative capacity of both goats’ organism and milk. Livest. Sci. 2018, 218, 37–43. [Google Scholar] [CrossRef]
- Christodoulou, C.; Kotsampasi, B.; Dotas, V.; Simoni, M.; Righi, F.; Tsiplakou, E. The effect of Spirulina supplementation in ewes’ oxidative status and milk quality. Anim. Feed Sci. Technol. 2023, 295, 115544. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Theodorou, G.; Politis, I.; Tsiplakou, E. Schizochytrium sp. Dietary supplementation modify Toll-like receptor 4 (TLR4) transcriptional regulation in monocytes and neutrophils of dairy goats. Cytokine 2021, 148, 155588. [Google Scholar] [CrossRef]
- Keller, M.; Manzocchi, E.; Rentsch, D.; Lugarà, R.; Giller, K. Antioxidant and inflammatory gene expression profiles of bovine peripheral blood mononuclear cells in response to arthrospira platensis before and after lps challenge. Antioxidants 2021, 10, 814. [Google Scholar] [CrossRef]
- Ognistaia, A.V.; Markina, Z.V.; Orlova, T.Y. Antimicrobial Activity of Marine Microalgae. Russ. J. Mar. Biol. 2022, 48, 217–230. [Google Scholar] [CrossRef]
- Tsiplakou, E.; Abdullah, M.A.M.; Skliros, D.; Chatzikonstantinou, M.; Flemetakis, E.; Labrou, N.; Zervas, G. The effect of dietary Chlorella vulgaris supplementation on micro-organism community, enzyme activities and fatty acid profile in the rumen liquid of goats. J. Anim. Physiol. Anim. Nutr. 2017, 101, 275–283. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Tsiplakou, E. The impact of the dietary supplementation level with Schizochytrium sp. on milk chemical composition and fatty acid profile, of both blood plasma and milk of goats. Small Rumin. Res. 2020, 193, 106252. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Skliros, D.; Sotirakoglou, K.; Flemetakis, E.; Tsiplakou, E. The effect of forage-to-concentrate ratio on schizochytrium spp.-supplemented goats: Modifying rumen microbiota. Animals 2021, 11, 2746. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Skliros, D.; Flemetakis, E.; Tsiplakou, E. Changes in the rumen bacteriome structure and enzymatic activities of goats in response to dietary supplementation with Schizochytrium spp. Microorganisms 2021, 9, 1528. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Sotirakoglou, K.; Skliros, D.; Flemetakis, E.; Tsiplakou, E. Dose and time response of dietary supplementation with Schizochytrium sp. on the abundances of several microorganisms in the rumen liquid of dairy goats. Livest. Sci. 2021, 247, 104489. [Google Scholar] [CrossRef]
- Mavrommatis, A.; Skliros, D.; Simoni, M.; Righi, F.; Flemetakis, E.; Tsiplakou, E. Alterations in the rumen particle-associated microbiota of goats in response to dietary supplementation levels of Schizochytrium spp. Sustainability 2021, 13, 607. [Google Scholar] [CrossRef]
- Musa, M.; Ayoko, G.A.; Ward, A.; Rösch, C.; Brown, R.J.; Rainey, T.J. Factors Affecting Microalgae Production for Biofuels and the Potentials of Chemometric Methods in Assessing and Optimizing Productivity. Cells 2019, 8, 851. [Google Scholar] [CrossRef] [Green Version]
- Wild, K.J.; Trautmann, A.; Katzenmeyer, M.; Steingaß, H.; Posten, C.; Rodehutscord, M. Chemical Composition and Nutritional Characteristics for Ruminants of the Microalgae Chlorella Vulgaris Obtained Using Different Cultivation Conditions. Algal Res. 2018, 38, 101385. [Google Scholar] [CrossRef]
- Costa, J.A.V.; Freitas, B.C.B.; Rosa, G.M.; Moraes, L.; Morais, M.G.; Mitchell, B.G. Operational and economic aspects of Spirulina-based biorefinery. Bioresour. Technol. 2019, 292, 121946. [Google Scholar] [CrossRef] [PubMed]
- Finamore, A.; Palmery, M.; Bensehaila, S.; Peluso, I. Antioxidant, Immunomodulating, and Microbial-Modulating Activities of the Sustainable and Ecofriendly Spirulina. Oxid. Med. Cell. Longev. 2017, 2017, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Christodoulou, C.; Mavrommatis, A.; Loukovitis, D.; Symeon, G.; Dotas, V.; Kotsampasi, B.; Tsiplakou, E. Inclusion of Camelina sativa Seeds in Ewes’ Diet Modifies Rumen Microbiota. Animals 2023, in press. [Google Scholar] [CrossRef]
- Mould, F.L.; Kliem, K.E.; Morgan, R.; Mauricio, R.M. In vitro microbial inoculum: A review of its function and properties. Anim. Feed Sci. Technol. 2005, 123–124 Pt 1, 31–50. [Google Scholar] [CrossRef]
- Ramos-Morales, E.; Arco-Pérez, A.; Martín-García, A.I.; Yáñez-Ruiz, D.R.; Frutos, P.; Hervás, G. Use of stomach tubing as an alternative to rumen cannulation to study ruminal fermentation and microbiota in sheep and goats. Anim. Feed Sci. Technol. 2014, 198, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Carberry, C.A.; Kenny, D.A.; Han, S.; McCabe, M.S.; Waters, S.M. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 2012, 78, 4949–4958. [Google Scholar] [CrossRef] [Green Version]
- Denman, S.E.; McSweeney, C.S. Development of a real-time PCR assay for monitoring anaerobic fungal and cellulolytic bacterial populations within the rumen. FEMS Microbiol. Ecol. 2006, 58, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, D.; Dubey, J.; Mehra, S. Probiotic efficiency of Spirulina platensis-stimulating growth of lactic acid bacteria. World J. Dairy Food Sci. 2009, 4, 160–163. [Google Scholar]
- Ozdemir, G.; Karabay, N.U.; Dalay, M.C.; Pazarbasi, B. Antibacterial activity of volatile component and various extracts of Spirulina platensis. Phytother. Res. 2004, 18, 754–757. [Google Scholar] [CrossRef]
- Balachandran, P.; Pugh, N.D.; Ma, G.; Pasco, D.S. Tolllike receptor 2-dependent activation of monocytes by Spirulina polysaccharide and its immune enhancing action in mice. Int. Immunopharmacol. 2006, 6, 1808–1814. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, P.; Chauhan, A. In vitro antibacterial activity of laboratory grown culture of Spirulina platensis. Indian J. Microbiol. 2008, 48, 348–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sheekh, M.M.; Daboo, S.; Swelim, M.A.; Mohamed, S. Production and characterization of antimicrobial active substance from Spirulina platensis. Iran. J. Microbiol. 2014, 6, 112–119. [Google Scholar]
- Beheshtipour, H.; Mortazavian, A.M.; Haratian, P.; Khosravi-Darani, K. Effects of Chlorella vulgaris and Arthrospira platensis addition on viability of probiotic bacteria in yogurt and its biochemical properties. Eur. Food Res. Technol. 2012, 235, 719–728. [Google Scholar] [CrossRef]
- Peluso, I.; Romanelli, L.; Palmery, M. Interactions between prebiotics, probiotics, polyunsaturated fatty acids and polyphenols: Diet or supplementation for metabolic syndrome prevention? Int. J. Food Sci. Nutr. 2014, 65, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Tapio, I.; Snelling, T.J.; Strozzi, F.; Wallace, R.J. The ruminal microbiome associated with methane emissions from ruminant livestock. J. Anim. Sci. Biotechnol. 2014, 8, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J. Anim. Sci. Biotechnol. 2017, 8, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Scheifinger, C.C.; Wolin, M.J. Propionate Formation from Cellulose and Soluble Sugars by Combined Cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 1973, 26, 789–795. [Google Scholar] [CrossRef]
- Tymensen, L.D.; Beauchemin, K.A.; McAllister, T.A. Structures of free-living and protozoa-associated methanogen communities in the bovine rumen differ according to comparative analysis of 16S rRNA and mcrA genes. Microbiology 2012, 158, 1808–1817. [Google Scholar] [CrossRef] [Green Version]
- Guyader, J.; Eugène, M.; Nozière, P.; Morgavi, D.P.; Doreau, M.; Martin, C. Influence of rumen protozoa on methane emission in ruminants: A meta-analysis approach. Animal 2014, 8, 1816–1825. [Google Scholar] [CrossRef]
- Newbold, C.J.; De la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The role of ciliate protozoa in the rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef] [Green Version]
- Abuelfatah, K.; Zuki, A.B.; Goh, Y.M.; Sazili, A.Q.; Abubakr, A. Effects of feeding whole linseed on ruminal fatty acid composition and microbial population in goats. Anim. Nutr. 2016, 2, 323–328. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Shinkai, T.; Koike, S. Ecological and physiological characterization shows that Fibrobacter succinogenes is important in rumen fiber digestion-Review. Folia Microbiol. 2008, 53, 195–200. [Google Scholar] [CrossRef]
- Harmon, D.L. Understanding starch utilization in the small intestine of cattle. Asian-Aust. J. Anim. Sci. 2009, 22, 915–922. [Google Scholar] [CrossRef]
- Owens, C.E.; Zinn, R.A.; Hassen, A.; Owens, F.N. Mathematical linkage of total-tract digestion of starch and neutral detergent fiber to their fecal concentrations and the effect of site of starch digestion on extent of digestion and energetic efficiency of cattle. Prof. Anim. Sci. 2016, 32, 531–549. [Google Scholar] [CrossRef] [Green Version]
- Brake, D.W.; Swanson, K.C. Ruminant nutrition symposium: Effects of postruminal flows of protein and amino acids on small intestinal starch digestion in beef cattle. J. Anim. Sci. 2018, 96, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Svihus, B.; Uhlen, A.K.; Harstad, O.M. Effect of starch granule structure, associated components and processing on nutritive value of cereal starch: A review. Anim. Feed Sci. Technol. 2005, 122, 303–320. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Krause, D.O.; Gozho, G.N.; McBride, B.W. Subacute ruminal acidosis in dairy cows: The physiological causes, incidence and consequences. Vet. J. 2008, 176, 21–31. [Google Scholar] [CrossRef]
- Freer, S.N. Purification and characterization of the extracellular α-amylase from Streptococcus bovis JB1. Appl. Environ. Microbiol. 1993, 59, 1398–1402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, Q.; Bird, S.H.; Gill, H.S.; Rowe, J.B. Immunological cross-reactivity between the vaccine and other isolates of Streptococcus bovis and Lactobacillus. FEMS Immunol. Med. Microbiol. 1999, 26, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asanuma, N.; Hino, T. Regulation of fermentation in a ruminal bacterium, Streptococcus bovis, with special reference to rumen acidosis. Anim. Sci. J. 2002, 73, 313–325. [Google Scholar] [CrossRef]
- Plaizier, J.C.; Mesgaran, M.D.; Derakhshani, H.; Golder, H.; Khafipour, E.; Kleen, J.L.; Lean, I.; Loor, J.; Penner, G.; Zebeli, Q. Review: Enhancing gastrointestinal health in dairy cows. Animals 2018, 12, S399–S418. [Google Scholar] [CrossRef] [Green Version]
- Neubauer, V.; Petri, R.M.; Humer, E.; Kröger, I.; Reisinger, N.; Baumgartner, W.; Wagner, M.; Zebeli, Q. Starch-rich diet induced rumen acidosis and hindgut dysbiosis in dairy cows of different lactations. Animals 2020, 10, 1727. [Google Scholar] [CrossRef]
- Gill, H.S.; Shu, Q.; Leng, R.A. Immunization with Streptococcus bovis protects against lactic acidosis in sheep. Vaccine 2000, 18, 2541–2548. [Google Scholar] [CrossRef] [PubMed]



| Dietary Treatments | Phyla (%) | |||
|---|---|---|---|---|
| Bacteroidetes | Firmicutes | Proteobacteria | Firmicutes:Bacteroidetes | |
| CON | 39.3 | 50.7 | 7.9 | 1.29 |
| SP5 | 26.5 | 59.4 | 12.5 | 2.24 |
| SP10 | 36.1 | 55.3 | 6.2 | 1.53 |
| SP15 | 29.2 | 63.0 | 5.8 | 2.16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Christodoulou, C.; Mavrommatis, A.; Loukovitis, D.; Symeon, G.; Dotas, V.; Kotsampasi, B.; Tsiplakou, E. Effect of Spirulina Dietary Supplementation in Modifying the Rumen Microbiota of Ewes. Animals 2023, 13, 740. https://doi.org/10.3390/ani13040740
Christodoulou C, Mavrommatis A, Loukovitis D, Symeon G, Dotas V, Kotsampasi B, Tsiplakou E. Effect of Spirulina Dietary Supplementation in Modifying the Rumen Microbiota of Ewes. Animals. 2023; 13(4):740. https://doi.org/10.3390/ani13040740
Chicago/Turabian StyleChristodoulou, Christos, Alexandros Mavrommatis, Dimitris Loukovitis, George Symeon, Vassilios Dotas, Basiliki Kotsampasi, and Eleni Tsiplakou. 2023. "Effect of Spirulina Dietary Supplementation in Modifying the Rumen Microbiota of Ewes" Animals 13, no. 4: 740. https://doi.org/10.3390/ani13040740








