Integrated Single-Trait and Multi-Trait GWASs Reveal the Genetic Architecture of Internal Organ Weight in Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Animal Samples and Phenotype Collection
2.3. Genotyping and Quality Control
2.4. Population Structure and Linkage Disequilibrium (LD) Estimation
2.5. Single-Trait and Multi-Trait Genome-Wide Association Studies
2.6. Estimation of Heritability and Phenotypic Variation
2.7. Candidate Gene Search and Function Analysis
3. Results and Discussion
3.1. Phenotype Statistics and Heritability Estimation
3.2. Population Structure and LD decay
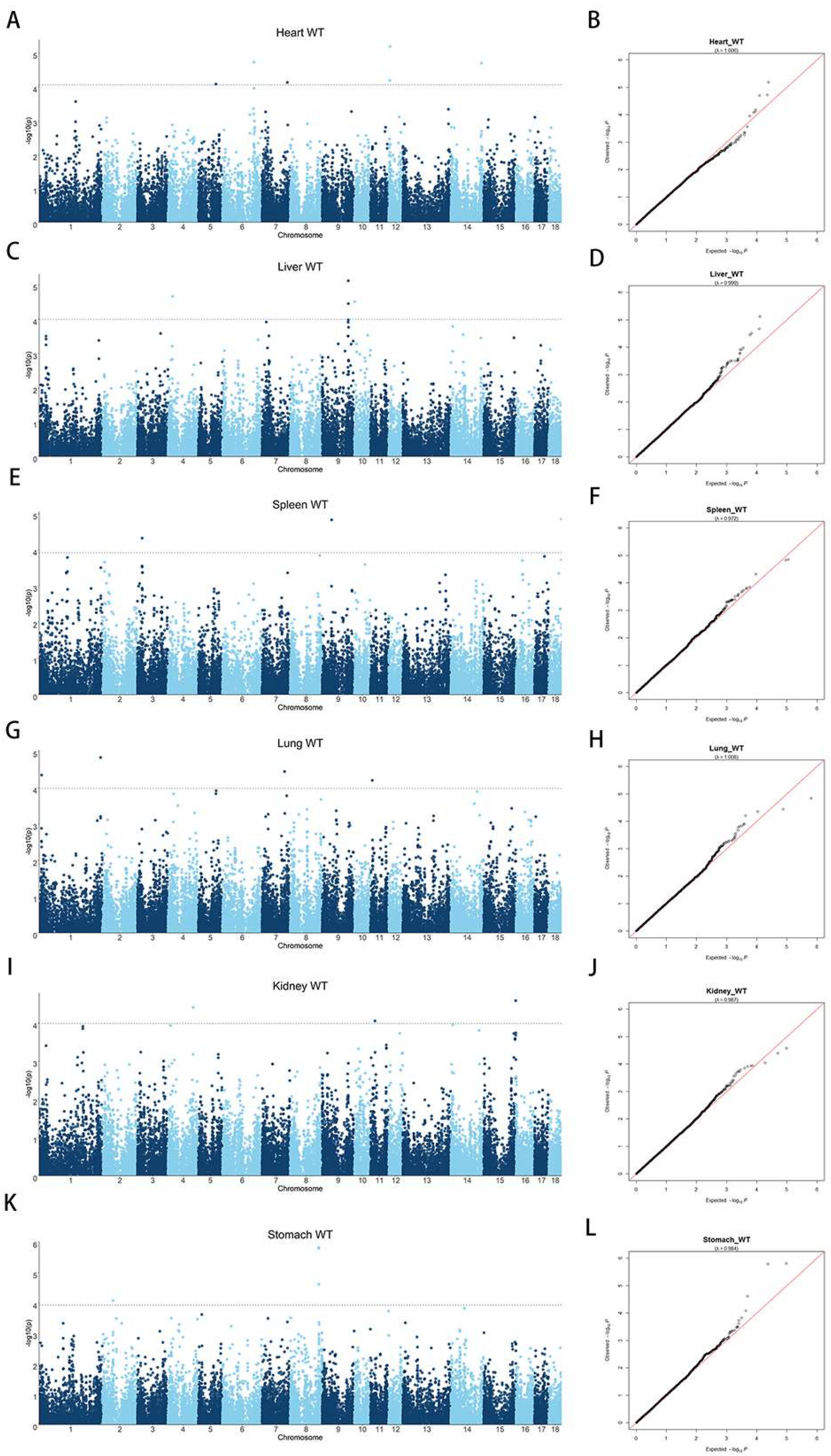
3.3. Single-Trait GWASs
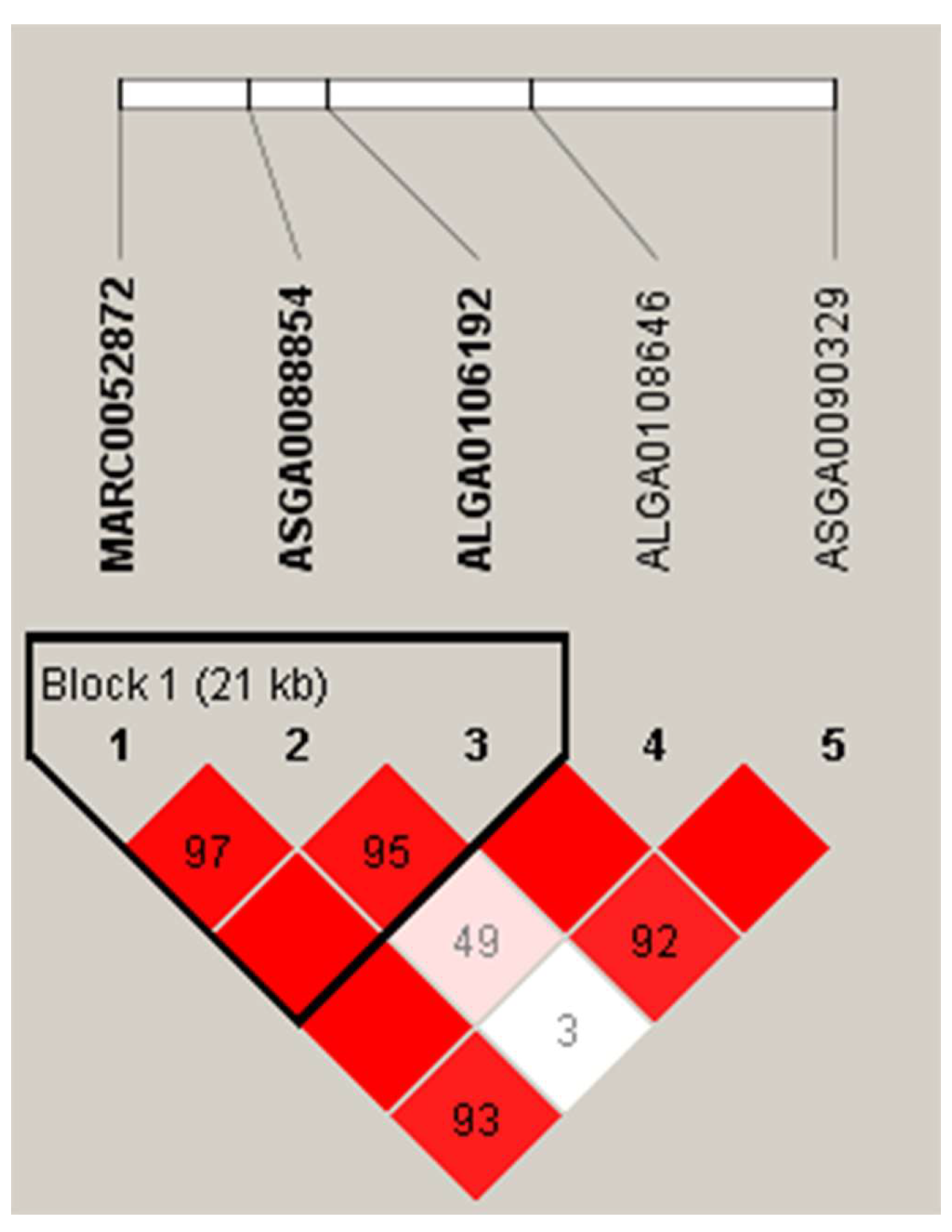
3.4. Haplotype Block Analysis
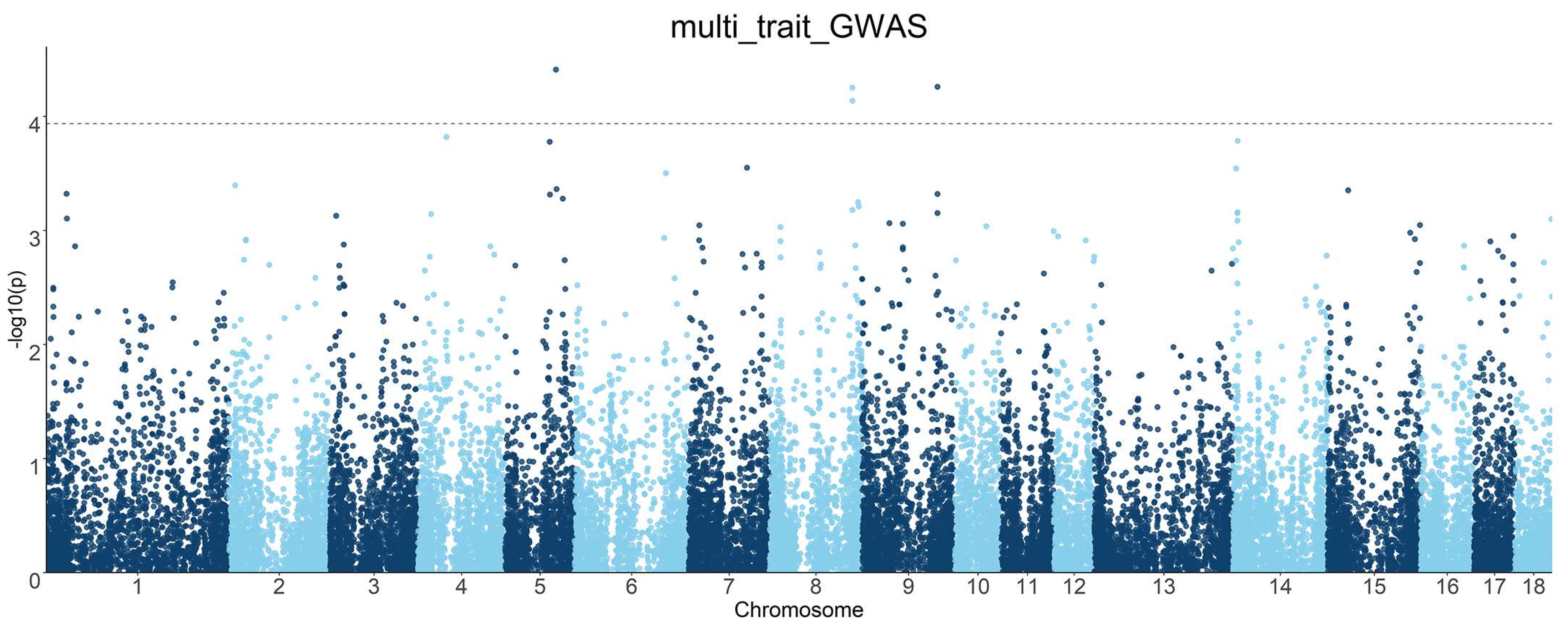
3.5. Multi-Trait GWASs
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- An, B.; Xia, J.; Chang, T.; Wang, X.; Miao, J.; Xu, L.; Zhang, L.; Gao, X.; Chen, Y.; Li, J.; et al. Genome-wide association study identifies loci and candidate genes for internal organ weights in Simmental beef cattle. Physiol. Genom. 2018, 50, 523–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mader, C.J.; Montanholi, Y.R.; Wang, Y.J.; Miller, S.P.; Mandell, I.B.; McBride, B.W.; Swanson, K.C. Relationships among measures of growth performance and efficiency with carcass traits, visceral organ mass, and pancreatic digestive enzymes in feedlot cattle. J. Anim. Sci. 2009, 87, 1548–1557. [Google Scholar] [CrossRef] [Green Version]
- Mubbunu, L.; Bowa, K.; Petrenko, V.; Silitongo, M. Correlation of Internal Organ Weights with Body Weight and Body Height in Normal Adult Zambians: A Case Study of Ndola Teaching Hospital. Anat. Res. Int. 2018, 2018, 4687538. [Google Scholar] [CrossRef]
- Hu, Z.L.; Park, C.A.; Reecy, J.M. Bringing the Animal QTLdb and CorrDB into the future: Meeting new challenges and providing updated services. Nucleic Acids Res. 2022, 50, D956–D961. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, X.; Zhang, F.; Su, Y.; Hou, L.; Chen, H.; Zhang, Z.; Huang, L. Multi-breed genome-wide association study reveals novel loci associated with the weight of internal organs. Genet. Sel. Evol. 2015, 47, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Xiong, Y.; Zuo, B.; Lei, M.; Jiang, S.; Li, F.; Zheng, R.; Li, J.; Xu, D. Detection of quantitative trait loci associated with several internal organ traits and teat number trait in a pig population. J. Genet. Genom. 2007, 34, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, L.; Liang, J. Genome-Wide Association Study for Certain Carcass Traits and Organ Weights in a Large White×Minzhu Intercross Porcine Population. J. Integr. Agric. 2014, 13, 2721–2730. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.H.; Duan, Y.; Haley, C.S.; Ren, J.; de Koning, D.J.; Huang, L.S. High throughput analyses of epistasis for swine body dimensions and organ weights. Anim. Genet. 2011, 42, 15–21. [Google Scholar] [CrossRef]
- Casale, F.P.; Rakitsch, B.; Lippert, C.; Stegle, O. Efficient set tests for the genetic analysis of correlated traits. Nat. Methods 2015, 12, 755–758. [Google Scholar] [CrossRef]
- Porter, H.F.; O’Reilly, P.F. Multivariate simulation framework reveals performance of multi-trait GWAS methods. Sci. Rep. 2017, 7, 38837. [Google Scholar] [CrossRef] [Green Version]
- Broadaway, K.A.; Cutler, D.J.; Duncan, R.; Moore, J.L.; Ware, E.B.; Jhun, M.A.; Bielak, L.F.; Zhao, W.; Smith, J.A.; Peyser, P.A.; et al. A Statistical Approach for Testing Cross-Phenotype Effects of Rare Variants. Am. J. Hum. Genet. 2016, 98, 525–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackinger, S.; Zeggini, E. Statistical methods to detect pleiotropy in human complex traits. Open Biol. 2017, 7, 170125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Ding, R.; Zhuang, Z.; Zeng, H.; Wen, S.; Ruan, D.; Wu, J.; Qiu, Y.; Zheng, E.; Cai, G.; et al. Genome-Wide Association Analysis Reveals Genetic Loci and Candidate Genes for Chest, Abdominal, and Waist Circumferences in Two Duroc Pig Populations. Front. Vet. Sci. 2021, 8, 807003. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Xu, L.; Xia, J.; Wang, X.; Miao, J.; Chang, T.; Song, M.; Ni, J.; Xu, L.; Zhang, L.; et al. Multiple association analysis of loci and candidate genes that regulate body size at three growth stages in Simmental beef cattle. BMC Genet. 2020, 21, 32. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Stephens, M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat. Methods 2014, 11, 407–409. [Google Scholar] [CrossRef]
- Yan, G.; Guo, T.; Xiao, S.; Zhang, F.; Xin, W.; Huang, T.; Xu, W.; Li, Y.; Zhang, Z.; Huang, L. Imputation-Based Whole-Genome Sequence Association Study Reveals Constant and Novel Loci for Hematological Traits in a Large-Scale Swine F(2) Resource Population. Front. Genet. 2018, 9, 401. [Google Scholar] [CrossRef] [Green Version]
- Glickman, M.E.; Rao, S.R.; Schultz, M.R. False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J. Clin. Epidemiol. 2014, 67, 850–857. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, R.; Quan, J.; Yang, M.; Wang, X.; Zheng, E.; Yang, H.; Fu, D.; Yang, Y.; Yang, L.; Li, Z.; et al. Genome-wide association analysis reveals genetic loci and candidate genes for feeding behavior and eating efficiency in Duroc boars. PLoS ONE 2017, 12, e0183244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaitlen, N.; Kraft, P. Heritability in the genome-wide association era. Hum. Genet. 2012, 131, 1655–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, Z.; Wu, J.; Xu, C.; Ruan, D.; Qiu, Y.; Zhou, S.; Ding, R.; Quan, J.; Yang, M.; Zheng, E.; et al. The Genetic Architecture of Meat Quality Traits in a Crossbred Commercial Pig Population. Foods 2022, 11, 3143. [Google Scholar] [CrossRef]
- Zhuang, Z.; Ding, R.; Peng, L.; Wu, J.; Ye, Y.; Zhou, S.; Wang, X.; Quan, J.; Zheng, E.; Cai, G.; et al. Genome-wide association analyses identify known and novel loci for teat number in Duroc pigs using single-locus and multi-locus models. BMC Genom. 2020, 21, 344. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef] [Green Version]
- Voss, O.H.; Tian, L.; Murakami, Y.; Coligan, J.E.; Krzewski, K. Emerging role of CD300 receptors in regulating myeloid cell efferocytosis. Mol. Cell. Oncol. 2015, 2, e964625. [Google Scholar] [CrossRef] [Green Version]
- Abplanalp, W.T.; Cremer, S.; John, D.; Hoffmann, J.; Schuhmacher, B.; Merten, M.; Rieger, M.A.; Vasa-Nicotera, M.; Zeiher, A.M.; Dimmeler, S. Clonal Hematopoiesis-Driver DNMT3A Mutations Alter Immune Cells in Heart Failure. Circ. Res. 2021, 128, 216–228. [Google Scholar] [CrossRef]
- Li, X.; Huang, Z.; Chen, Y.; Sun, X.; Yi, Z.; Xie, J.; Yu, X.; Chen, H.; Zhong, J. Case report of two affected siblings in a family with thiamine metabolism dysfunction syndrome 5: A rare, but treatable neurodegenerative disease. BMC Neurol. 2022, 22, 373. [Google Scholar] [CrossRef]
- Giacometti, R.; Kronberg, F.; Biondi, R.M.; Passeron, S. Catalytic isoforms Tpk1 and Tpk2 of Candida albicans PKA have non-redundant roles in stress response and glycogen storage. Yeast 2009, 26, 273–285. [Google Scholar] [CrossRef] [Green Version]
- Di Renzo, F.; Doneda, L.; Menegola, E.; Sardella, M.; De Vecchi, G.; Collini, P.; Spreafico, F.; Fossati-Bellani, F.; Giavini, E.; Radice, P.; et al. The murine Pou6f2 gene is temporally and spatially regulated during kidney embryogenesis and its human homolog is overexpressed in a subset of Wilms tumors. J. Pediatr. Hematol. Oncol. 2006, 28, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Zhu, H.; Xu, J.; Wang, Y.; Zhang, Y.; Zhang, M.; Zhu, D. Long non-coding RNA POU6F2-AS2 promotes cell proliferation and drug resistance in colon cancer by regulating miR-377/BRD4. J. Cell. Mol. Med. 2020, 24, 4136–4149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Song, X.; Zhao, Y. circNBPF10/miR-224 Axis Regulates PBX3 to Promote the Malignant Progression of Lung Cancer. J. Oncol. 2022, 2022, 2832920. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, H.; Yang, Y.; Kang, L. Long Noncoding RNA Urothelial Carcinoma-Associated 1 Promotes the Proliferation and Metastasis of Human Lung Tumor Cells by Regulating MicroRNA-144. Oncol. Res. 2018, 26, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Yang, T.; Huang, H.; Alarmanazi, F.; Liu, G. Uncoupling of UNC5C with Polymerized TUBB3 in Microtubules Mediates Netrin-1 Repulsion. J. Neurosci. 2017, 37, 5620–5633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyamoto, Y.; Futamura, M.; Kitamura, N.; Nakamura, Y.; Baba, H.; Arakawa, H. Identification of UNC5A as a novel transcriptional target of tumor suppressor p53 and a regulator of apoptosis. Int. J. Oncol. 2010, 36, 1253–1260. [Google Scholar] [CrossRef] [Green Version]
- Danesh, S.M.; Villasenor, A.; Chong, D.; Soukup, C.; Cleaver, O. BMP and BMP receptor expression during murine organogenesis. Gene Expr. Patterns 2009, 9, 255–265. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhao, X.Y.; Zhu, J.S.; Chen, N.W.; Fan, H.N.; Yang, W.; Guo, J.H. CCR7 regulates ANO6 to promote migration of pancreatic ductal adenocarcinoma cells via the ERK signaling pathway. Oncol. Lett. 2018, 16, 2599–2605. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Qiu, H.; Xiao, S.; Wu, Z.; Yang, M.; Yang, J.; Ren, J.; Huang, L. A genome-wide association study identifies genomic loci associated with backfat thickness, carcass weight, and body weight in two commercial pig populations. J. Appl. Genet. 2017, 58, 499–508. [Google Scholar] [CrossRef]

| Trait | N | Mean (±SD) | Min | Max | C.V.% a | h2 (±SE) |
|---|---|---|---|---|---|---|
| Heart WT | 1518 | 455.57 ± 78.06 | 217.1 | 868.8 | 17.13 | 0.21 ± 0.04 |
| Liver WT | 1518 | 1763.61 ± 271.83 | 993.9 | 2607.7 | 15.41 | 0.46 ± 0.04 |
| Spleen WT | 1517 | 212.54 ± 47.49 | 95.9 | 502.1 | 22.34 | 0.49 ± 0.04 |
| Lung WT | 1517 | 1020.54 ± 240.10 | 918.9 | 2090.4 | 2.18 | 0.28 ± 0.04 |
| Kidney WT | 1486 | 0.41 ± 0.08 | 0.17 | 0.84 | 19.51 | 0.36 ± 0.04 |
| Stomach WT | 1518 | 727.68 ± 129.97 | 495.7 | 1321.7 | 17.86 | 0.47 ± 0.04 |
| Heart WT | Liver WT | Spleen WT | Lung WT | Kidney WT | Stomach WT | |
|---|---|---|---|---|---|---|
| Heart WT | 1 | 0.36 | 0.33 | 0.41 | 0.37 | 0.49 |
| Liver WT | 0.31 ± 0.11 | 1 | 0.34 | −0.02 | 0.62 | 0.38 |
| Spleen WT | 0.23 ± 0.11 | 0.11 ± 0.09 | 1 | 0.15 | 0.33 | 0.41 |
| Lung WT | 0.33 ± 0.13 | 0.17 ± 0.11 | 0.04 ± 0.11 | 1 | 0.05 | 0.27 |
| Kidney WT | 0.30 ± 012 | 0.33 ± 0.09 | 0.07 ± 0.10 | 0.29 ± 0.12 | 1 | 0.43 |
| Stomach WT | 0.02 ± 0.12 | 0.03 ± 0.09 | 0.26 ± 0.09 | 0.02 ± 0.11 | 0.03 ± 0.10 | 1 |
| Trait | SSC | SNP | Position (bp) | MAF | p-Value | PEV (%) a | Candidate Gene | Distance |
|---|---|---|---|---|---|---|---|---|
| Heart WT | 6 | WU_10.2_6_126961053 | 137,008,535 | 0.257 | 1.88 × 10−5 | 2.09% | ST6GALNAC3 | Within |
| 14 | 6_43731895 | 132,228,312 | 0.407 | 1.99 × 10−5 | 1.71% | HTRA1 | 124,717 | |
| 12 | WU_10.2_12_6703865 | 6,682,110 | 0.389 | 6.40 × 10−5 | 1.70% | CD300LB | 44,143 | |
| 5 | ALGA0032998 | 76,317,972 | 0.264 | 8.30 × 10−5 | 1.36% | ANO6 | Within | |
| 7 | WU_10.2_7_116585612 | 110,088,932 | 0.242 | 7.42 × 10−5 | 1.10% | KCNK10 | −29,135 | |
| 12 | 12_5381300 | 5,426,010 | 0.319 | 6.50 × 10−5 | 0.53% | CDK3 | Within | |
| Liver WT | 4 | WU_10.2_4_20570494 | 19,550,555 | 0.249 | 2.15 × 10−5 | 2.11% | CCN3 | −35,758 |
| 9 | H3GA0028070 | 113,152,352 | 0.09 | 7.54 × 10−5 | 2.10% | TPK1 | Within | |
| 9 | ASGA0044340 | 113,140,343 | 0.118 | 3.51 × 10−5 | 0.82% | TPK1 | Within | |
| 10 | WU_10.2_10_3469625 | 1,753,591 | 0.476 | 3.08 × 10−5 | 0.43% | RGS21 | Within | |
| Spleen WT | 18 | ALGA0098928 | 54,993,603 | 0.328 | 1.39 × 10−5 | 2.22% | POU6F2 | Within |
| 3 | ALGA0105765 | 20,525,651 | 0.216 | 4.74 × 10−5 | 0.78% | HS3ST4 | −19,582 | |
| 9 | WU_10.2_9_45824613 | 40,989,995 | 0.399 | 1.47 × 10−5 | 0.58% | TTC12 | Within | |
| Lung WT | 11 | ALGA0060656 | 8,759,687 | 0.231 | 6.24 × 10−5 | 2.76% | FRY | Within |
| 1 | ALGA0110225 | 266,708,292 | 0.278 | 1.46 × 10−5 | 1.81% | PBX3 | 97,646 | |
| 7 | ASGA0035515 | 98,022,168 | 0.04 | 3.57 × 10−5 | 1.43% | YLPM1 | Within | |
| 1 | MARC0089438 | 11,012,474 | 0.271 | 4.46 × 10−5 | 0.69% | / | / | |
| Kidney WT | 15 | WU_10.2_15_153747936 | 138,955,316 | 0.369 | 2.61 × 10−5 | 1.97% | / | / |
| 4 | WU_10.2_4_119054114 | 108,872,194 | 0.478 | 3.93 × 10−5 | 1.62% | RAP1A | 122,469 | |
| 11 | WU_10.2_11_20231427 | 19,917,312 | 0.221 | 8.86 × 10−5 | 0.88% | SUCLA2 | Within | |
| Stomach WT | 8 | ALGA0106192 | 124,443,641 | 0.064 | 1.54 × 10−5 | 2.62% | UNC5C | Within |
| 8 | MARC0052872 | 124,421,940 | 0.063 | 1.61 × 10−5 | 2.53% | UNC5C | Within | |
| 8 | ASGA0101191 | 124,548,240 | 0.081 | 2.40 × 10−5 | 2.14% | BMPR1B | Within | |
| 2 | MARC0018316 | 46,014,239 | 0.46 | 8.35 × 10−5 | 0.74% | ARNTL | Within |
| SSC | SNP | Position (bp) | MAF | p-Value | PEV (%) a | Candidate Gene | Distance |
|---|---|---|---|---|---|---|---|
| 8 | ALGA0106192 | 124,443,641 | 0.064 | 7.29 × 10−5 | 2.62% | UNC5C | Within |
| 8 | MARC0052872 | 124,421,940 | 0.063 | 5.61 × 10−5 | 2.53% | UNC5C | Within |
| 9 | H3GA0028070 | 113,152,352 | 0.088 | 5.51 × 10−5 | 2.10% | TPK1 | Within |
| 5 | ALGA0032998 | 76,317,972 | 0.266 | 3.88 × 10−5 | 1.36% | ANO6 | Within |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wu, J.; Zhuang, Z.; Ye, Y.; Zhou, S.; Qiu, Y.; Ruan, D.; Wang, S.; Yang, J.; Wu, Z.; et al. Integrated Single-Trait and Multi-Trait GWASs Reveal the Genetic Architecture of Internal Organ Weight in Pigs. Animals 2023, 13, 808. https://doi.org/10.3390/ani13050808
Li X, Wu J, Zhuang Z, Ye Y, Zhou S, Qiu Y, Ruan D, Wang S, Yang J, Wu Z, et al. Integrated Single-Trait and Multi-Trait GWASs Reveal the Genetic Architecture of Internal Organ Weight in Pigs. Animals. 2023; 13(5):808. https://doi.org/10.3390/ani13050808
Chicago/Turabian StyleLi, Xuehua, Jie Wu, Zhanwei Zhuang, Yong Ye, Shenping Zhou, Yibin Qiu, Donglin Ruan, Shiyuan Wang, Jie Yang, Zhenfang Wu, and et al. 2023. "Integrated Single-Trait and Multi-Trait GWASs Reveal the Genetic Architecture of Internal Organ Weight in Pigs" Animals 13, no. 5: 808. https://doi.org/10.3390/ani13050808
APA StyleLi, X., Wu, J., Zhuang, Z., Ye, Y., Zhou, S., Qiu, Y., Ruan, D., Wang, S., Yang, J., Wu, Z., Cai, G., & Zheng, E. (2023). Integrated Single-Trait and Multi-Trait GWASs Reveal the Genetic Architecture of Internal Organ Weight in Pigs. Animals, 13(5), 808. https://doi.org/10.3390/ani13050808





