First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Sample Collection
2.2. DNA Extraction and RCA
2.3. PCR Screening and Recovery of Full-Length Sequences
2.4. Data Analysis
3. Results and Discussion
3.1. Fecal Samples
3.1.1. Isolation of BMMF Molecules from Feces
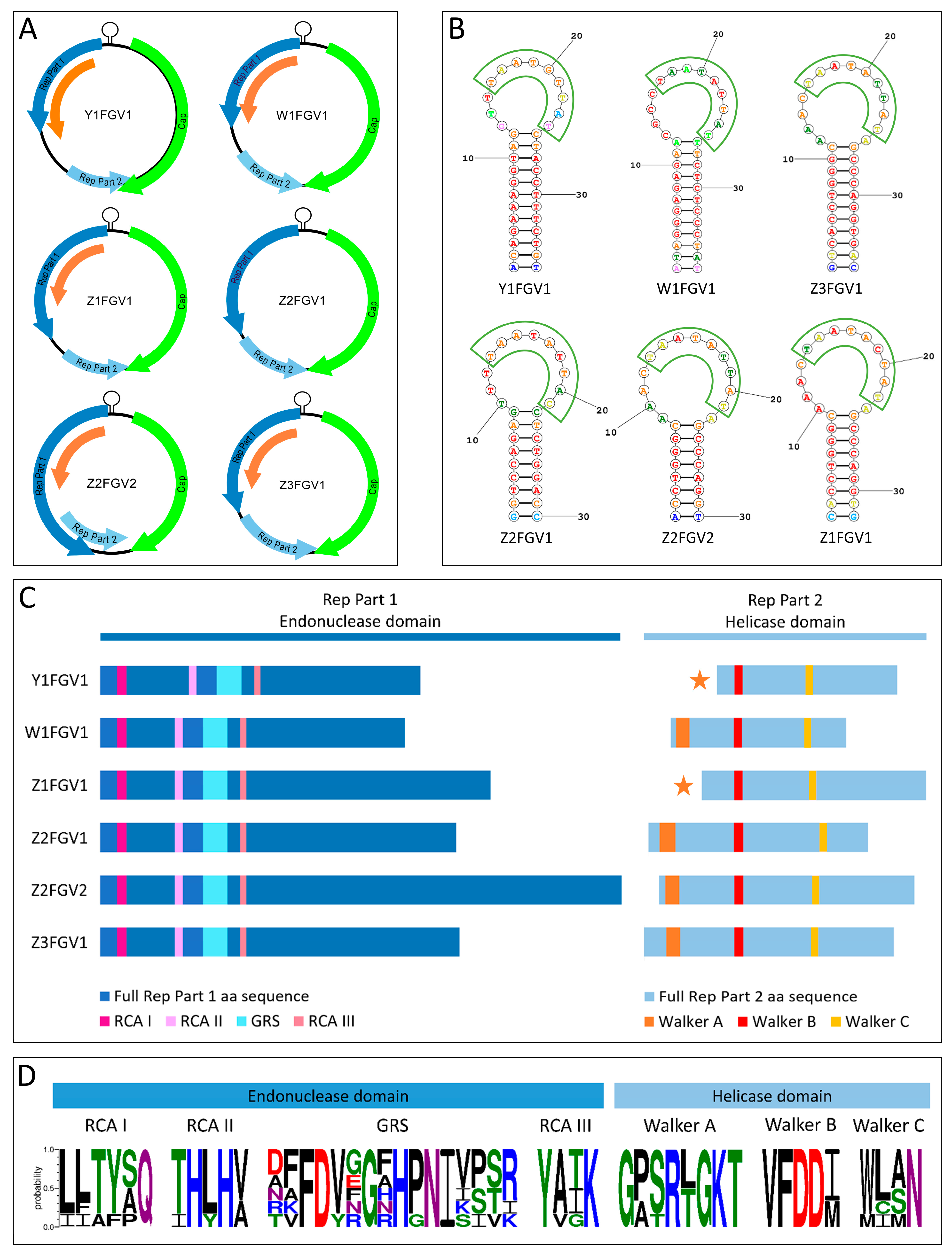
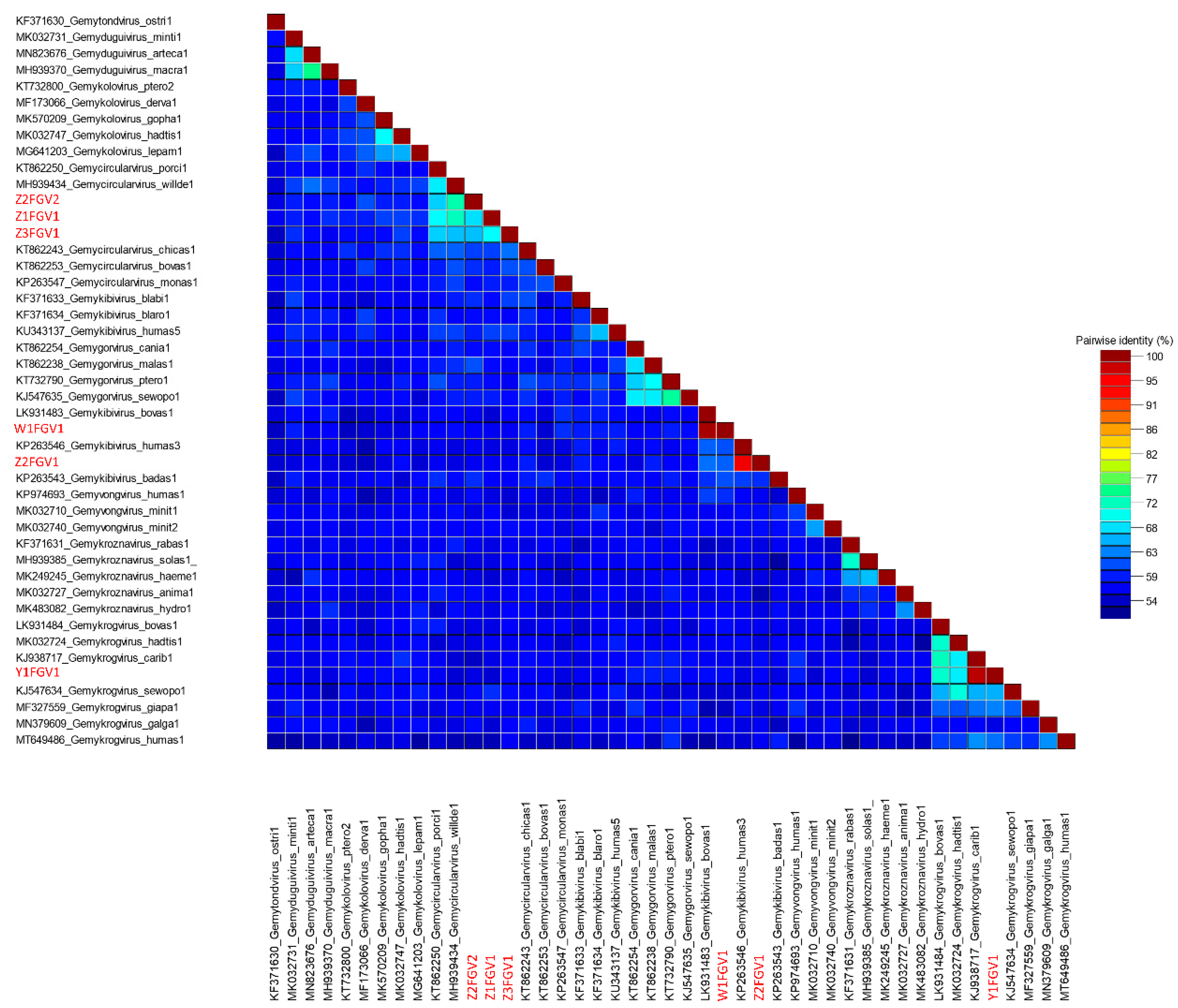
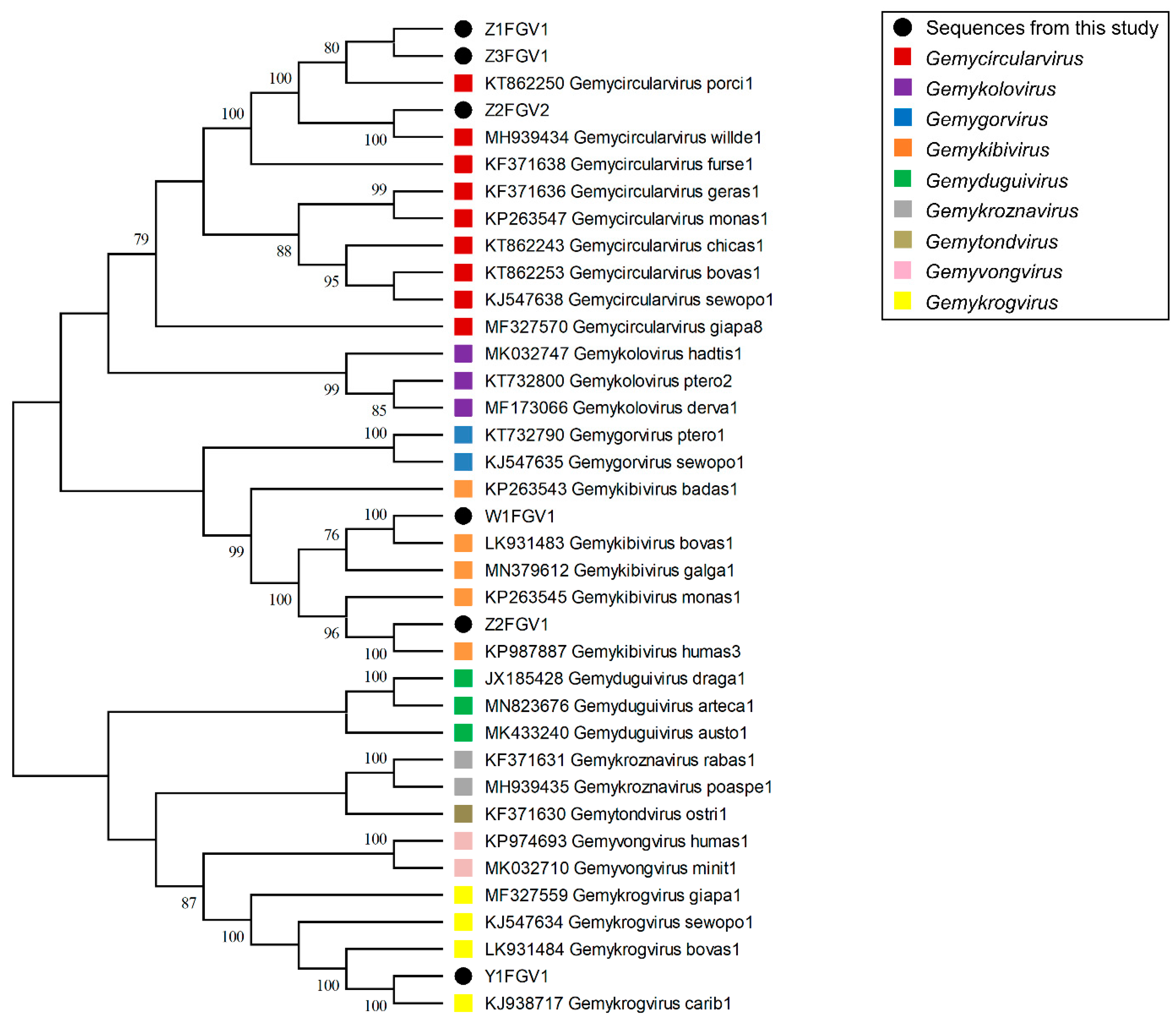
3.1.2. Isolation of CRESS DNA Viruses from Feces
3.2. Blood and Serum Samples
3.3. Matched Blood and Fecal Samples
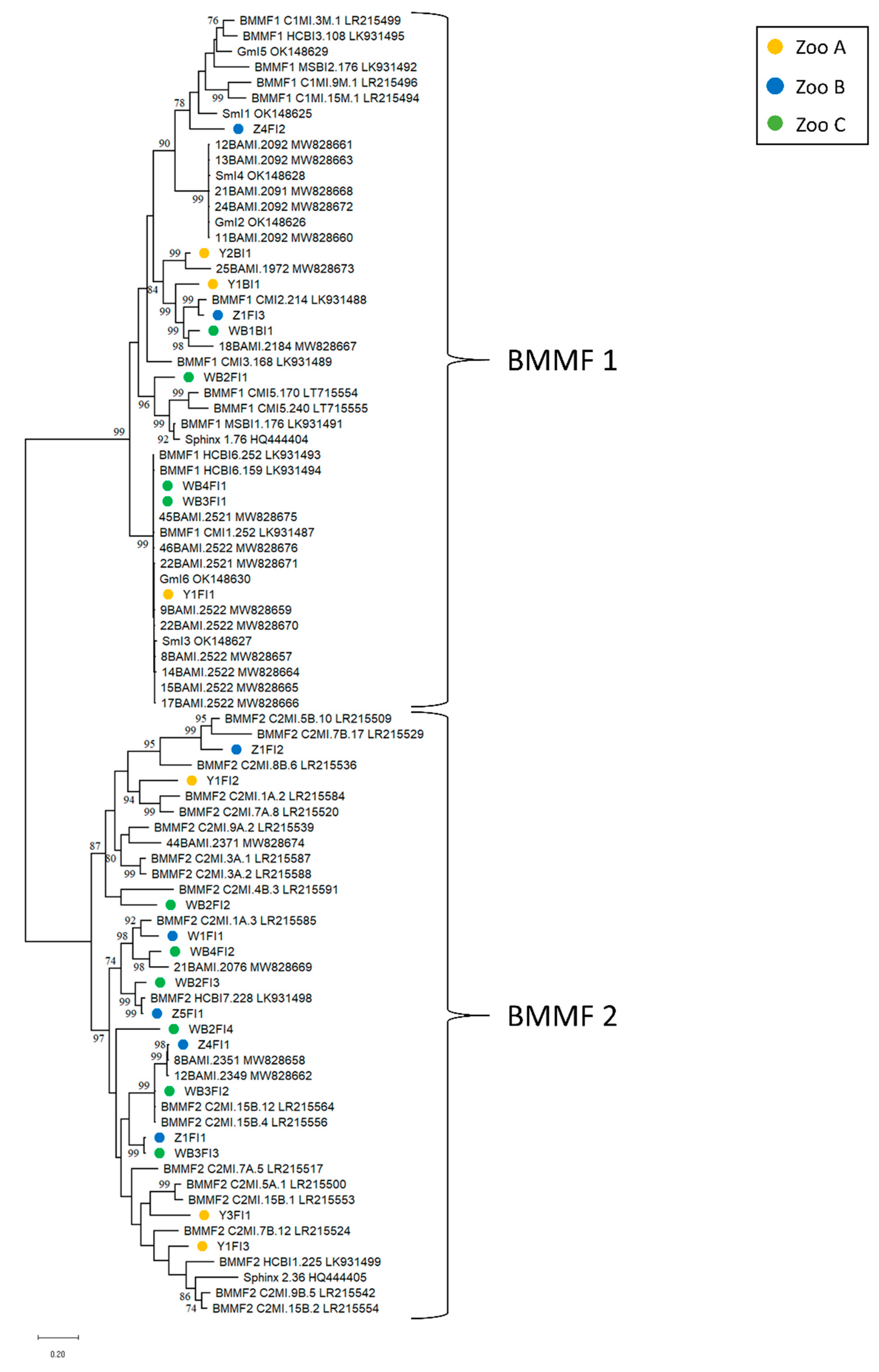
3.3.1. BMMF in Blood and Feces
3.3.2. CRESS DNA Viruses in Blood and Feces
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faria, F.; Filho, A.; Madalena, F.; Josahkian, L. Pedigree analysis in the Brazilian Zebu breeds. J. Anim. Breed. Genet. 2009, 126, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Leslie, D.M., Jr.; Schaller, G.B. Bos grunniens and Bos mutus (Artiodactyla: Bovidae). Mamm. Species 2009, 836, 1–17. [Google Scholar] [CrossRef]
- Kugonza, D.; Nabasirye, M.; Mpairwe, D.; Hanotte, O.; Okeyo, A. Productivity and morphology of Ankole cattle in three livestock production systems in Uganda. Anim. Genet. Resour./Resour. Génétiques Anim./Recur. Genéticos Anim. 2011, 48, 13–22. [Google Scholar] [CrossRef]
- Pasha, T.; Hayat, Z. Present situation and future perspective of buffalo production in Asia. J. Anim. Plant Sci. 2012, 22, 250–256. [Google Scholar]
- de Villiers, E.M.; Gunst, K.; Chakraborty, D.; Ernst, C.; Bund, T.; zur Hausen, H. A specific class of infectious agents isolated from bovine serum and dairy products and peritumoral colon cancer tissue. Emerg. Microbes Infect. 2019, 8, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H.; Bund, T.; de Villiers, E.M. Infectious Agents in Bovine Red Meat and Milk and Their Potential Role in Cancer and Other Chronic Diseases. In Viruses, Genes, and Cancer; Hunter, E., Bister, K., Compans, R.W., Eds.; Springer: Cham, Switzerland, 2017; Volume 407, pp. 83–116. [Google Scholar]
- Manuelidis, L. Nuclease resistant circular DNAs copurify with infectivity in scrapie and CJD. J. Neurovirol. 2011, 17, 131–145. [Google Scholar] [CrossRef]
- Lamberto, I.; Gunst, K.; Muller, H.; zur Hausen, H.; de Villiers, E.M. Mycovirus-like DNA virus sequences from cattle serum and human brain and serum samples from multiple sclerosis patients. Genome Announc. 2014, 2, e00848-14. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol. 2017, 3, vew037. [Google Scholar] [CrossRef]
- Gunst, K.; zur Hausen, H.; de Villiers, E.M. Isolation of bacterial plasmid-related replication-associated circular DNA from a serum sample of a multiple sclerosis patient. Genome Announc. 2014, 2, e00847-14. [Google Scholar] [CrossRef]
- Whitley, C.; Gunst, K.; Muller, H.; Funk, M.; zur Hausen, H.; de Villiers, E.M. Novel replication-competent circular DNA molecules from healthy cattle serum and milk and multiple sclerosis-affected human brain tissue. Genome Announc. 2014, 2, e00849-14. [Google Scholar] [CrossRef]
- Falida, K.; Eilebrecht, S.; Gunst, K.; zur Hausen, H.; de Villiers, E.M. Isolation of Two Virus-Like Circular DNAs from Commercially Available Milk Samples. Genome Announc. 2017, 5, e00266-17. [Google Scholar] [CrossRef]
- Gilbert, W.; Dressler, D. DNA replication: The rolling circle model. Cold Spring Harb. Symp. Quant. Biol. 1968, 33, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Dressler, D.; Wolfson, J. The Rolling Circle for ϕX DNA Replication, III. Synthesis of Supercoiled Duplex Rings. Proc. Natl. Acad. Sci. 1970, 67, 456–463. [Google Scholar] [CrossRef] [PubMed]
- Baas, P.D. DNA replication of single-stranded Escherichia coli DNA phages. Biochim. Et Biophys. Acta (BBA)—Gene Struct. Expr. 1985, 825, 111–139. [Google Scholar] [CrossRef]
- Koonin, E.V.; Ilyina, T.V. Geminivirus replication proteins are related to prokaryotic plasmid rolling circle DNA replication initiator proteins. J. Gen. Virol. 1992, 73, 2763–2766. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Masó, J.A.; MachóN, C.; Bordanaba-Ruiseco, L.; Espinosa, M.; Coll, M.; Del Solar, G. Plasmid Rolling-Circle Replication. Microbiol. Spectr. 2015, 3, 45–69. [Google Scholar] [CrossRef]
- Krupovic, M.; Varsani, A.; Kazlauskas, D.; Breitbart, M.; Delwart, E.; Rosario, K.; Yutin, N.; Wolf, Y.I.; Harrach, B.; Zerbini, F.M.; et al. Cressdnaviricota: A Virus Phylum Unifying Seven Families of Rep-Encoding Viruses with Single-Stranded, Circular DNA Genomes. J. Virol. 2020, 94, e00582-20. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Family Genomoviridae: 2021 taxonomy update. Arch. Virol. 2021, 166, 2911–2926. [Google Scholar] [CrossRef]
- Sikorski, A.; Massaro, M.; Kraberger, S.; Young, L.M.; Smalley, D.; Martin, D.P.; Varsani, A. Novel myco-like DNA viruses discovered in the faecal matter of various animals. Virus Res. 2013, 177, 209–216. [Google Scholar] [CrossRef]
- Levy, H.; Fontenele, R.S.; Harding, C.; Suazo, C.; Kraberger, S.; Schmidlin, K.; Djurhuus, A.; Black, C.E.; Hart, T.; Smith, A.L.; et al. Identification and Distribution of Novel Cressdnaviruses and Circular Molecules in Four Penguin Species in South Georgia and the Antarctic Peninsula. Viruses 2020, 12, 1029. [Google Scholar] [CrossRef]
- Orton, J.P.; Morales, M.; Fontenele, R.S.; Schmidlin, K.; Kraberger, S.; Leavitt, D.J.; Webster, T.H.; Wilson, M.A.; Kusumi, K.; Dolby, G.A. Virus discovery in desert tortoise fecal samples: Novel circular single-stranded DNA viruses. Viruses 2020, 12, 143. [Google Scholar] [CrossRef] [PubMed]
- Tisza, M.J.; Pastrana, D.V.; Welch, N.L.; Stewart, B.; Peretti, A.; Starrett, G.J.; Pang, Y.S.; Krishnamurthy, S.R.; Pesavento, P.A.; McDermott, D.H.; et al. Discovery of several thousand highly diverse circular DNA viruses. eLife 2020, 9, e51971. [Google Scholar] [CrossRef]
- Cibulski, S.; Alves de Lima, D.; Fernandes Dos Santos, H.; Teixeira, T.F.; Tochetto, C.; Mayer, F.Q.; Roehe, P.M. A plate of viruses: Viral metagenomics of supermarket chicken, pork and beef from Brazil. Virology 2021, 552, 1–9. [Google Scholar] [CrossRef]
- Wiederkehr, M.A.; Qi, W.; Schoenbaechler, K.; Fraefel, C.; Kubacki, J. Virus Diversity, Abundance, and Evolution in Three Different Bat Colonies in Switzerland. Viruses 2022, 14, 1911. [Google Scholar] [CrossRef]
- Custer, J.M.; White, R.; Taylor, H.; Schmidlin, K.; Fontenele, R.S.; Stainton, D.; Kraberger, S.; Briskie, J.V.; Varsani, A. Diverse single-stranded DNA viruses identified in New Zealand (Aotearoa) South Island robin (Petroica australis) fecal samples. Virology 2022, 565, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Krupovic, M.; Ghabrial, S.A.; Jiang, D.; Varsani, A. Genomoviridae: A new family of widespread single-stranded DNA viruses. Arch. Virol. 2016, 161, 2633–2643. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Varsani, A.; Koonin, E.V.; Krupovic, M. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat. Commun. 2019, 10, 3425. [Google Scholar] [CrossRef] [PubMed]
- Funk, M.; Gunst, K.; Lucansky, V.; Muller, H.; zur Hausen, H.; de Villiers, E.M. Isolation of protein-associated circular DNA from healthy cattle serum. Genome Announc. 2014, 2, e00846-14. [Google Scholar] [CrossRef]
- zur Hausen, H.; Bund, T.; de Villiers, E.M. Specific nutritional infections early in life as risk factors for human colon and breast cancers several decades later. Int. J. Cancer 2018, 144, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- Bund, T.; Nikitina, E.; Chakraborty, D.; Ernst, C.; Gunst, K.; Boneva, B.; Tessmer, C.; Volk, N.; Brobeil, A.; Weber, A.; et al. Analysis of chronic inflammatory lesions of the colon for BMMF Rep antigen expression and CD68 macrophage interactions. Proc. Natl. Acad. Sci. USA 2021, 118, e2025830118. [Google Scholar] [CrossRef]
- de Villiers, E.M.; zur Hausen, H. Bovine Meat and Milk Factors (BMMFs): Their Proposed Role in Common Human Cancers and Type 2 Diabetes Mellitus. Cancers 2021, 13, 5407. [Google Scholar] [CrossRef]
- Nikitina, E.; Alikhanyan, K.; Neßling, M.; Richter, K.; Kaden, S.; Ernst, C.; Seitz, S.; Chuprikova, L.; Häfele, L.; Gunst, K. Structural Expression of BMMF in tissues of colorectal, lung and pancreatic cancer patients. Int. J. Cancer 2022, 1–10. [Google Scholar] [CrossRef]
- Nikitina, E.; Burk-Körner, A.; Wiesenfarth, M.; Alwers, E.; Heide, D.; Tessmer, C.; Ernst, C.; Krunic, D.; Schrotz-King, P.; Chang-Claude, J. Bovine meat and milk factor protein expression in tumor-free mucosa of colorectal cancer patients coincides with macrophages and might interfere with patient survival. Mol. Oncol. 2023. [Google Scholar] [CrossRef] [PubMed]
- König, M.-T.; Fux, R.; Link, E.; Sutter, G.; Märtlbauer, E.; Didier, A. Circular Rep-Encoding Single-Stranded DNA Sequences in Milk from Water Buffaloes (Bubalus arnee f. bubalis). Viruses 2021, 13, 1088. [Google Scholar] [CrossRef] [PubMed]
- König, M.-T.; Fux, R.; Link, E.; Sutter, G.; Märtlbauer, E.; Didier, A. Identification and Characterization of Circular Single-Stranded DNA Genomes in Sheep and Goat Milk. Viruses 2021, 13, 2176. [Google Scholar] [CrossRef]
- Lechmann, J.; Ackermann, M.; Kaiser, V.; Bachofen, C. Viral infections shared between water buffaloes and small ruminants in Switzerland. J. Vet. Diagn. Investig. 2021, 33, 894–905. [Google Scholar] [CrossRef]
- Pohl, S.; Habermann, D.; Link, E.K.; Fux, R.; Boldt, C.L.; Franz, C.M.A.P.; Hölzel, C.; Klempt, M. Detection of DNA sequences attributed to bovine meat and milk factors (BMMF/SPHINX) in food-related samples. Food Control 2022, 135, 108779. [Google Scholar] [CrossRef]
- Scherf, B.; Pilling, D. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; FAO: Rome, Italy, 2015; ISBN 978-92-5-108820-3. [Google Scholar]
- Zhang, K.; Lenstra, J.A.; Zhang, S.; Liu, W.; Liu, J. Evolution and domestication of the Bovini species. Anim. Genet. 2020, 51, 637–657. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Burland, T.G. DNASTAR’s Lasergene Sequence Analysis Software. In Bioinformatics Methods and Protocols; Misener, S., Krawetz, S.A., Eds.; Humana Press: Totowa, NJ, USA, 2000; Volume 132, pp. 71–91. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A Virus Classification Tool Based on Pairwise Sequence Alignment and Identity Calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- The European Molecular Biology Open Software Suite (EMBOSS) A. Available online: http://emboss.bioinformatics.nl/cgi-bin/emboss/equicktandem (accessed on 10 March 2023).
- The European Molecular Biology Open Software Suite (EMBOSS) B. Available online: https://www.bioinformatics.nl/cgi-bin/emboss/palindrome (accessed on 10 March 2023).
- National Center for Biotechnology Information (NCBI). ORFfinder. Available online: https://www.ncbi.nlm.nih.gov/orffinder/ (accessed on 17 March 2023).
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Rosario, K.; Mettel, K.A.; Benner, B.E.; Johnson, R.; Scott, C.; Yusseff-Vanegas, S.Z.; Baker, C.C.M.; Cassill, D.L.; Storer, C.; Varsani, A.; et al. Virus discovery in all three major lineages of terrestrial arthropods highlights the diversity of single-stranded DNA viruses associated with invertebrates. PeerJ 2018, 6, e5761. [Google Scholar] [CrossRef]
- Zhao, L.; Rosario, K.; Breitbart, M.; Duffy, S. Eukaryotic Circular Rep-Encoding Single-Stranded DNA (CRESS DNA) Viruses: Ubiquitous Viruses With Small Genomes and a Diverse Host Range. Adv. Virus Res. 2019, 103, 71–133. [Google Scholar] [CrossRef] [PubMed]
- Ling, Y.; Zhang, X.; Qi, G.; Yang, S.; Jingjiao, L.; Shen, Q.; Wang, X.; Cui, L.; Hua, X.; Deng, X.; et al. Viral metagenomics reveals significant viruses in the genital tract of apparently healthy dairy cows. Arch. Virol. 2019, 164, 1059–1067. [Google Scholar] [CrossRef]
- Kraberger, S.; Waits, K.; Ivan, J.; Newkirk, E.; VandeWoude, S.; Varsani, A. Identification of circular single-stranded DNA viruses in faecal samples of Canada lynx (Lynx canadensis), moose (Alces alces) and snowshoe hare (Lepus americanus) inhabiting the Colorado San Juan Mountains. Infect. Genet. Evol. 2018, 64, 1–8. [Google Scholar] [CrossRef]
- Khalifeh, A.; Blumstein, D.T.; Fontenele, R.S.; Schmidlin, K.; Richet, C.; Kraberger, S.; Varsani, A. Diverse cressdnaviruses and an anellovirus identified in the fecal samples of yellow-bellied marmots. Virology 2021, 554, 89–96. [Google Scholar] [CrossRef]
- Kraberger, S.; Argüello-Astorga, G.R.; Greenfield, L.G.; Galilee, C.; Law, D.; Martin, D.P.; Varsani, A. Characterisation of a diverse range of circular replication-associated protein encoding DNA viruses recovered from a sewage treatment oxidation pond. Infect. Genet. Evol. 2015, 31, 73–86. [Google Scholar] [CrossRef]
- Conceição-Neto, N.; Zeller, M.; Heylen, E.; Lefrère, H.; Mesquita, J.R.; Matthijnssens, J. Fecal virome analysis of three carnivores reveals a novel nodavirus and multiple gemycircularviruses. Virol. J. 2015, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Ng, T.F.F.; Chen, L.-F.; Zhou, Y.; Shapiro, B.; Stiller, M.; Heintzman, P.D.; Varsani, A.; Kondov, N.O.; Wong, W.; Deng, X. Preservation of viral genomes in 700-y-old caribou feces from a subarctic ice patch. Proc. Natl. Acad. Sci. USA 2014, 111, 16842–16847. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, S.; Shan, T.; Hou, R.; Liu, Z.; Li, W.; Guo, L.; Wang, Y.; Chen, P.; Wang, X. Virome comparisons in wild-diseased and healthy captive giant pandas. Microbiome 2017, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Fontenele, R.S.; Lacorte, C.; Lamas, N.S.; Schmidlin, K.; Varsani, A.; Ribeiro, S.G. Single stranded DNA viruses associated with capybara faeces sampled in Brazil. Viruses 2019, 11, 710. [Google Scholar] [CrossRef]
- Fontenele, R.S.; Roumagnac, P.; Richet, C.; Kraberger, S.; Stainton, D.; Aleamotu‘a, M.; Filloux, D.; Bernardo, P.; Harkins, G.W.; McCarthy, J. Diverse genomoviruses representing twenty-nine species identified associated with plants. Arch. Virol. 2020, 165, 2891–2901. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Zhao, M.; Wang, H.; Hou, R.; Qin, K.; Qian, Y.; Zhang, H.; Zhou, Y.; Wu, W.; Gu, J. Virome of Giant Panda-Infesting Ticks Reveals Novel Bunyaviruses and Other Viruses That Are Genetically Close to Those from Giant Pandas. Microbiol. Spectr. 2022, 10, e02034-22. [Google Scholar] [CrossRef]
- Waits, K.; Edwards, M.J.; Cobb, I.N.; Fontenele, R.S.; Varsani, A. Identification of an anellovirus and genomoviruses in ixodid ticks. Virus Genes 2018, 54, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Dayaram, A.; Marinov, M.; Ware, J.; Kraberger, S.; Stainton, D.; Breitbart, M.; Varsani, A. Diverse circular ssDNA viruses discovered in dragonflies (Odonata: Epiprocta). J. Gen. Virol. 2012, 93, 2668–2681. [Google Scholar] [CrossRef]
- Singh, G.; Ramamoorthy, S. Potential for the cross-species transmission of swine torque teno viruses. Vet. Microbiol. 2018, 215, 66–70. [Google Scholar] [CrossRef]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef]

| Species/Breed | Serum | EDTA-Blood | Feces | No. of Individuals |
|---|---|---|---|---|
| Watusi | 22 | 6 | 4 * | 28 |
| Zebu | 13 | 10 | 6 * | 23 |
| Yak | 1 | 4 | 2 * | 5 |
| Water buffalo | - | 4 | 3 * + 1 | 5 |
| Individual | Species/Breed | Zoo/Origin | Isolated Molecules | Length in Nt | Acc. No. | Sample Material | DNA-Type |
|---|---|---|---|---|---|---|---|
| Y1 | Yak | A | Y1FI1 | 2522 | OQ633422 | Feces | BMMF1 |
| Y1FI2 | 2387 | OQ633423 | Feces | BMMF2 | |||
| Y1FI3 | 2346 | OQ633424 | Feces | BMMF2 | |||
| Y1FGV1 | 2232 | OQ633425 | Feces | Genomoviridae | |||
| Y1BI1 | 1616 | OQ633426 | EDTA-blood | BMMF1 | |||
| Y2 | Yak | A | Y2BI1 | 1959 | OQ633427 | EDTA-blood | BMMF1 |
| Y3 | Yak | A | Y3FI1 | 2229 | OQ633428 | Feces | BMMF2 |
| W1 | Watusi | B | W1FI1 | 2238 | OQ633429 | Feces | BMMF2 |
| W1FGV1 | 2134 | OQ633430 | Feces | Genomoviridae | |||
| Z1 | Zebu | B | Z1FI1 | 2297 | OQ633431 | Feces | BMMF2 |
| Z1FI2 | 2733 | OQ633432 | Feces | BMMF2 | |||
| Z1FI3 | 2146 | OQ633433 | Feces | BMMF1 | |||
| Z1FGV1 | 2215 | OQ633434 | Feces | Genomoviridae | |||
| Z2 | Zebu | B | Z2FGV1 | 2118 | OQ633435 | Feces | Genomoviridae |
| Z2FGV2 | 2216 | OQ633436 | Feces | Genomoviridae | |||
| Z3 | Zebu | B | Z3FGV1 | 2200 | OQ633437 | Feces | Genomoviridae |
| Z4 | Zebu | B | Z4FI1 | 2353 | OQ633438 | Feces | BMMF2 |
| Z4FI2 | 2118 | OQ633439 | Feces | BMMF1 | |||
| Z5 | Zebu | B | Z5FI1 | 2280 | OQ633440 | Feces | BMMF2 |
| WB1 | Water Buffalo | C | WB1BI1 | 2185 | OQ633441 | EDTA-blood | BMMF1 |
| WB2 | Water Buffalo | C | WB2FI1 | 1913 | OQ633442 | Feces | BMMF1 |
| WB2FI2 | 2476 | OQ633443 | Feces | BMMF2 | |||
| WB2FI3 | 2375 | OQ633444 | Feces | BMMF2 | |||
| WB2FI4 | 2447 | OQ633445 | Feces | BMMF2 | |||
| WB3 | Water Buffalo | C | WB3FI1 | 2521 | OQ633446 | Feces | BMMF1 |
| WB3FI2 | 2359 | OQ633447 | Feces | BMMF2 | |||
| WB3FI3 | 2298 | OQ633448 | Feces | BMMF2 | |||
| WB4 | Water Buffalo | C | WB4FI1 | 2522 | OQ633449 | Feces | BMMF1 |
| WB4FI2 | 2380 | OQ633450 | Feces | BMMF2 |
| Isolate | Period Size | TR | IR | Nt between TR and IR |
|---|---|---|---|---|
| Y1FI1 | 22 | ATACCCCTACGTTTACCGATCA | TAAATGCTTTTA | 50 |
| Z1FI3 | 22 | ATACTCCTAGGTTTACCTACCA | TAAATGCTTTTA | 50 |
| Z4FI2 | 22 | CTACGTTTACCCATCAATACCC | TAAATGCTTTTA | 56 |
| WB2FI1 | 22 | CCTACGTTTACCGATCAATACC | TAAATGCTTTTA | 55 |
| WB3FI1 | 22 | ATACCCCTACGTTTACCGATCA | TAAATGCTTTTA | 50 |
| WB4FI1 | 22 | ATACCCCTACGTTTACCGATCA | TAAATGCTTTTA | 50 |
| Y1BI1 | 22 | TACCAATACTCCTAGGTTTACC | TAAATGCTTTTA | 53 |
| Y2BI1 | 22 | CACCGTTTACCCATCAATATGA | TAAATGCTTTTA | 56 |
| WB1BI1 | 22 | ATACTCCTAGGTTTACCTACCA | TAAATGCTTTTA | 50 |
| Isolate | Nonanucleotide | Motif I | Motif II | GRS Motif | Motif III | Walker A | Walker B | Walker C |
|---|---|---|---|---|---|---|---|---|
| Y1FGV1 | TAATATTAT | IITFPQ | IHYHV | TAFDYFGAHGNIKSVR | YVGK | GPTRTGKT | VFDDI | MCMN |
| Z1FGV1 | TAATACTAT | LLAYAQ | THLHV | DFFDVGGHHPNIVPSR | YATK | GASRLGKT | VFDDM | WLAN |
| Z2FGV2 | TAATATTAT | LLTYSQ | THLHV | DFFDVNGNHPNIVPSR | YATK | GPSRLGKT | VFDDI | WLAN |
| Z3FGV1 | TAATATTAT | LLTYSQ | THLHV | NFFDVRGRHPNIVPSR | YAIK | GASRLGKT | VFDDI | WLAN |
| W1FGV1 | TAATGTTAT | LLTYAQ | THLHA | AVFDVGGFHPNISITK | YAIK | GPSRTGKT | VFDDI | WISN |
| Z2FGV1 | TAATATTAC | LFTYSQ | THLHA | RKFDVEGFHPNIISTI | YATK | GPSRTGKT | VFDDM | WLSN |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
König, M.-T.; Frölich, K.; Jandowsky, A.; Knauf-Witzens, T.; Langner, C.; Dietrich, R.; Märtlbauer, E.; Didier, A. First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle. Animals 2023, 13, 1492. https://doi.org/10.3390/ani13091492
König M-T, Frölich K, Jandowsky A, Knauf-Witzens T, Langner C, Dietrich R, Märtlbauer E, Didier A. First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle. Animals. 2023; 13(9):1492. https://doi.org/10.3390/ani13091492
Chicago/Turabian StyleKönig, Marie-Thérèse, Kai Frölich, Anabell Jandowsky, Tobias Knauf-Witzens, Christoph Langner, Richard Dietrich, Erwin Märtlbauer, and Andrea Didier. 2023. "First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle" Animals 13, no. 9: 1492. https://doi.org/10.3390/ani13091492
APA StyleKönig, M.-T., Frölich, K., Jandowsky, A., Knauf-Witzens, T., Langner, C., Dietrich, R., Märtlbauer, E., & Didier, A. (2023). First Insights into the Occurrence of Circular Single-Stranded DNA Genomes in Asian and African Cattle. Animals, 13(9), 1492. https://doi.org/10.3390/ani13091492





