Genomic Characterizations of Porcine Epidemic Diarrhea Viruses (PEDV) in Diarrheic Piglets and Clinically Healthy Adult Pigs from 2019 to 2022 in China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Sample Information
2.2. Real-Time PCR Detection and Genomic Sequencing
2.3. Sequence Alignment, Recombination and Phylogenetic Analyses
3. Results
3.1. Chinese PEDV Strains from 2019 to 2022 from Different Ages of Pigs Obtained New Genetic Characteristics
3.2. Genomic Characterization Identified Novel PEDV Recombinants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Have, P.; Moving, V.; Svansson, V.; Uttenthal, Å.; Bloch, B. Coronavirus infection in mink (Mustela vision). Serological evidence of infection with a coronavirus related to transmissible gastroenteritis virus and porcine epidemic diarrhea virus. Veter. Microbiol. 1992, 31, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Kang, B.-K.; Lee, C.-S.; Song, D.-S. Impact of porcine group A rotavirus co-infection on porcine epidemic diarrhea virus pathogenicity in piglets. Res. Veter. Sci. 2008, 84, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Fang, L.; Ding, Z.; Wang, D.; Peng, G.; Xiao, S. Rapid manipulation of the porcine epidemic diarrhea virus genome by CRISPR/Cas9 technology. J. Virol. Methods 2019, 276, 113772. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Dickerman, A.W.; Piñeyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.-J. Origin, Evolution, and Genotyping of Emergent Porcine Epidemic Diarrhea Virus Strains in the United States. mBio 2013, 4, e00737-13. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar]
- Smoľak, D.; Šalamúnová, S.; Jacková, A.; Haršányová, M.; Budiš, J.; Szemes, T.; Vilček, Š. Analysis of RNA virome in rectal swabs of healthy and diarrheic pigs of different age. Comp. Immunol. Microbiol. Infect. Dis. 2022, 90–91, 101892. [Google Scholar] [CrossRef]
- Van Reeth, K.; Pensaert, M. Prevalence of infections with enzootic respiratory and enteric viruses in feeder pigs entering fattening herds. Veter. Rec. 1994, 135, 594–597. [Google Scholar]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.; Culhane, M.R.; Rossow, K.D.; Rovira, A.; Collins, J.; Saif, L.J. Distinct Characteristics and Complex Evolution of PEDV Strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, L.; Shang, Y.; Tan, R.; Ji, M.; Yue, X.; Wang, N.; Liu, J.; Wang, C.; Li, Y.; et al. Emergence and evolution of highly pathogenic porcine epidemic diarrhea virus by natural recombination of a low pathogenic vaccine isolate and a highly pathogenic strain in the spike gene. Virus Evol. 2020, 6, veaa049. [Google Scholar]
- Qiu, M.; Li, S.; Xiao, Y.; Li, J.; Zhang, Y.; Li, X.; Feng, B.; Li, C.; Lin, H.; Zhu, J.; et al. Pathogenic and metagenomic evaluations reveal the correlations of porcine epidemic diarrhea virus, porcine kobuvirus and porcine astroviruses with neonatal piglet diarrhea. Microb. Pathog. 2022, 170, 105703. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-H.; Bae, J.-L.; Kang, T.-J.; Kim, J.; Chung, G.-H.; Lim, C.-W.; Laude, H.; Yang, M.-S.; Jang, Y.-S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells 2002, 14, 295–299. [Google Scholar] [PubMed]
- Cruz, D.J.M.; Kim, C.-J.; Shin, H.-J. Phage-displayed peptides having antigenic similarities with porcine epidemic diarrhea virus (PEDV) neutralizing epitopes. Virology 2006, 354, 28–34. [Google Scholar] [CrossRef]
- Sun, D.; Feng, L.; Shi, H.; Chen, J.; Cui, X.; Chen, H.; Liu, S.; Tong, Y.; Wang, Y.; Tong, G. Identification of two novel B cell epitopes on porcine epidemic diarrhea virus spike protein. Vet. Microbiol. 2008, 131, 73–81. [Google Scholar] [CrossRef]
- Chen, B.; Dong, S.; Yu, L.; Si, F.; Li, C.; Xie, C.; Yu, R.; Li, Z. Three Amino Acid Substitutions in the Spike Protein Enable the Coronavirus Porcine Epidemic Diarrhea Virus to Infect Vero Cells. Microbiol. Spectr. 2023, 11, e03872-22. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Lin, C.-M.; Yokoyama, M.; Yount, B.L.; Marthaler, D.; Douglas, A.L.; Ghimire, S.; Qin, Y.; Baric, R.S.; Saif, L.J.; et al. Deletion of a 197-Amino-Acid Region in the N-Terminal Domain of Spike Protein Attenuates Porcine Epidemic Diarrhea Virus in Piglets. J. Virol. 2017, 91, e00227-17. [Google Scholar] [CrossRef]
- Li, Z.; Ma, Z.; Dong, L.; Yang, T.; Li, Y.; Jiao, D.; Han, W.; Zheng, H.; Xiao, S. Molecular Mechanism of Porcine Epidemic Diarrhea Virus Cell Tropism. mBio 2022, 13, e00227-17. [Google Scholar] [CrossRef]
- Suzuki, T.; Terada, Y.; Enjuanes, L.; Ohashi, S.; Kamitani, W. S1 Subunit of Spike Protein from a Current Highly Virulent Porcine Epidemic Diarrhea Virus Is an Important Determinant of Virulence in Piglets. Viruses 2018, 10, 467. [Google Scholar] [CrossRef]
- Zhang, H.; Han, F.; Shu, X.; Li, Q.; Ding, Q.; Hao, C.; Yan, X.; Xu, M.; Hu, H. Co-infection of porcine epidemic diarrhoea virus and porcine deltacoronavirus enhances the disease severity in piglets. Transbound. Emerg. Dis. 2021, 69, 1715–1726. [Google Scholar] [CrossRef]
- Gerdts, V.; Zakhartchouk, A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2016, 206, 45–51. [Google Scholar] [CrossRef]
- Niu, X.; Wang, Q. Prevention and Control of Porcine Epidemic Diarrhea: The Development of Recombination-Resistant Live Attenuated Vaccines. Viruses 2022, 14, 1317. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Enjuanes, L.; Ziebuhr, J.; Snijder, E.J. Nidovirales: Evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Li, S.; Zhou, R.; Zhu, M.; He, S.; Ye, M.; Huang, Y.; Li, S.; Zhu, C.; Xia, P.; et al. Two novel porcine epidemic diarrhea virus (PEDV) recombinants from a natural recombinant and distinct subtypes of PEDV variants. Virus Res. 2017, 242, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Fang, L.; Ye, X.; Chen, J.; Xu, S.; Zhu, X.; Miao, Y.; Wang, D.; Xiao, S. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound Emerg. Dis. 2019, 66, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, Y.; Liu, Y.; Chen, Y.; Jiao, W.; Feng, H.; Wei, Q.; Wang, J.; Zhang, Y.; Zhang, G. Isolation and Identification of a Recombinant Porcine Epidemic Diarrhea Virus with a Novel Insertion in S1 Domain. Front. Microbiol. 2021, 12, 667084. [Google Scholar] [CrossRef]
- Sun, R.-Q.; Cai, R.-J.; Chen, Y.-Q.; Liang, P.-S.; Chen, D.-K.; Song, C.-X. Outbreak of Porcine Epidemic Diarrhea in Suckling Piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef]
- Chen, N.; Li, S.; Tian, Y.; Li, X.; Li, S.; Li, J.; Qiu, M.; Sun, Z.; Xiao, Y.; Yan, X.; et al. Chimeric HP-PRRSV2 containing an ORF2-6 consensus sequence induces antibodies with broadly neutralizing activity and confers cross protection against virulent NADC30-like isolate. Vet. Res. 2021, 52, 74. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef]
- Chen, N.; Ye, M.; Li, S.; Huang, Y.; Zhou, R.; Yu, X.; Tian, K.; Zhu, J. Emergence of a novel highly pathogenic recombinant virus from three lineages of porcine reproductive and respiratory syndrome virus 2 in China 2017. Transbound. Emerg. Dis. 2018, 65, 1775–1785. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef]
- Turlewicz-Podbielska, H.; Pomorska-Mól, M. Porcine Coronaviruses: Overview of the State of the Art. Virol. Sin. 2021, 36, 833–851. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zheng, Z.; Wang, H.; Yi, S.; Zhang, G.; Gong, L. The New Porcine Epidemic Diarrhea Virus Outbreak May Mean That Existing Commercial Vaccines Are Not Enough to Fully Protect against the Epidemic Strains. Front. Veter. Sci. 2021, 8, 697839. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Q.; Zhao, R.; Liu, J.; Zhao, Q.; Zhu, L.; Zhang, B.; Bi, J.; Yang, G.; Liu, J.; Yin, G. Epidemiology and phylogeny of spike gene of porcine epidemic diarrhea virus from Yunnan, China. Virus Res. 2018, 249, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fang, L.; Xiao, S. Porcine epidemic diarrhea in China. Virus Res. 2016, 226, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Zambrano-Moreno, C.; Pineda, P.; Calderón, C.; Rincón-Monroy, M.A.; Diaz, A.; Marthaler, D.G. Several lineages of porcine epidemic diarrhea virus in Colombia during the 2014 and 2016 epidemic. Transbound. Emerg. Dis. 2020, 68, 2465–2476. [Google Scholar] [CrossRef]
- Hanke, D.; Pohlmann, A.; Sauter-Louis, C.; Höper, D.; Stadler, J.; Ritzmann, M.; Steinrigl, A.; Schwarz, B.-A.; Akimkin, V.; Fux, R.; et al. Porcine Epidemic Diarrhea in Europe: In-Detail Analyses of Disease Dynamics and Molecular Epidemiology. Viruses 2017, 9, 177. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Murakami, S.; Takahashi, O.; Kodera, A.; Masuda, T.; Itoh, S.; Miyazaki, A.; Ohashi, S.; Tsutsui, T. Molecular characterization of pig epidemic diarrhoea viruses isolated in Japan from 2013 to 2014. Infect. Genet. Evol. 2015, 36, 363–368. [Google Scholar] [CrossRef]
- Vui, D.T.; Thanh, T.L.; Tung, N.; Srijangwad, A.; Tripipat, T.; Chuanasa, T.; Nilubol, D. Complete genome characterization of porcine epidemic diarrhea virus in Vietnam. Arch. Virol. 2015, 160, 1931–1938. [Google Scholar] [CrossRef]
- Li, Y.; Liang, J.; Wu, S.; Yan, Z.; Zhang, W. Complete genomic sequence analysis and intestinal tissue localization of a porcine Kobuvirus variant in China. Infect. Genet. Evol. 2022, 104, 105362. [Google Scholar] [CrossRef]


| Year/Location | Sample No. | PEDV Positive No. 1 | PEDV-Positive Percentages |
|---|---|---|---|
| Year | |||
| 2019 | 55 | 28 | 50.91% |
| 2020 | 150 | 16 | 10.67% |
| 2021 | 9 | 1 | 11.11% |
| 2022 | 118 | 4 | 3.39% |
| Total | 332 | 49 | 14.76% |
| Location | |||
| Jiangsu | 70 | 4 | 5.71% |
| Xinjiang | 39 | 28 | 71.79% |
| Guangdong | 23 | 0 | 0% |
| Henan | 84 | 5 | 5.95% |
| Shandong | 71 | 9 | 12.68% |
| Fujian | 45 | 3 | 6.67% |
| Total | 332 | 49 | 14.76% |
| Age | |||
| <10 days old | 58 | 29 | 50.00% |
| Adult | 274 | 20 | 7.30% |
| Total | 332 | 49 | 14.76% |
| No. | Name | Region (City, Province) | Date | Sample | Age | Symptom |
|---|---|---|---|---|---|---|
| 1 | XJ1904-700 | Kashi, Xinjiang | April 2019 | Intestine | 5 days old | Diarrhea, death |
| 2 | JSNJ2004-919 | Nanjing, Jiangsu | April 2020 | Intestine | Adult pig | No enteric disease |
| 3 | FJFZ2004-1017 | Fuzhou, Fujian | April 2020 | Intestine | Adult pig | No enteric diseases |
| 4 | JSNT2112-1248 | Nantong, Jiangsu | December 2021 | Intestine | 2 days old | Diarrhea, death |
| 5 | HNZMD2202-1405 | Zhumadian, Henan | February 2022 | Intestine | Adult pig | No enteric diseases |
| Gene/Genome | 5′UTR | ORF1ab | S | ORF3 | E | M | N | 3′UTR | Complete Genome | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Encoded Protein | - | pp1ab | Spike Protein | Hypothetical Protein 3 | Envelope Protein | Membrane Protein | Nucleocapsid Protein | - | - | |||||||
| nt 1 | nt | aa 2 | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | nt | ||
| Identity (%) to CV777 | XJ1904-700 | 97.60 3 | 97.08 | 97.54 | 92.96 | 92.14 | - | - | 96.10 | 96.05 | 97.94 | 97.79 | 96.83 | 97.73 | 97.90 | 96.30 |
| JSNJ2004-919 | 98.97 | 97.08 | 97.60 | 94.82 | 94.14 | - | - | 94.81 | 96.05 | 97.65 | 96.90 | 95.93 | 96.83 | 98.20 | 96.55 4 | |
| FJFZ2004-1017 | 98.97 | 97.30 | 97.85 | 93.16 | 92.50 | - | - | 95.67 | 96.05 | 97.50 | 97.35 | 95.55 | 96.60 | 96.44 | 96.43 | |
| JSNT2112-1248 | 98.29 | 97.15 | 97.77 | 93.30 | 92.43 | - | - | 95.24 | 96.05 | 97.06 | 96.90 | 95.93 | 96.83 | 98.20 | 96.36 | |
| HNZMD2202-1405 | 98.29 | 97.21 | 97.94 | 93.32 | 92.36 | - | - | 94.81 | 94.74 | 97.21 | 96.90 | 95.85 | 96.60 | 98.50 | 96.41 | |
| Identity (%) to AJ1102 | XJ1904-700 | 98.29 | 97.93 | 98.42 | 97.72 | 98.05 | 99.11 | 100 | 99.57 | 100 | 99.41 | 100 | 97.74 | 97.96 | 99.10 | 97.96 |
| JSNJ2004-919 | 99.66 | 98.55 | 98.75 | 94.62 | 94.52 | 96.15 | 97.32 | 98.27 | 100 | 98.53 | 98.23 | 95.93 | 97.05 | 98.80 | 97.77 | |
| FJFZ2004-1017 | 99.66 | 98.82 | 98.94 | 97.26 | 98.20 | 96.15 | 97.32 | 99.13 | 100 | 98.38 | 98.67 | 96.15 | 96.83 | 97.03 | 98.35 | |
| JSNT2112-1248 | 99.66 | 98.57 | 98.89 | 97.31 | 98.05 | 96.00 | 97.32 | 98.70 | 100 | 97.94 | 98.23 | 96.08 | 97.05 | 98.80 | 98.17 | |
| HNZMD2202-1405 | 99.66 | 98.68 | 99.10 | 97.48 | 98.05 | 96.15 | 97.32 | 98.27 | 98.68 | 98.09 | 98.23 | 96.00 | 96.83 | 99.10 | 98.29 | |
| Identity (%) to LW/L | XJ1904-700 | 98.63 | 97.87 | 98.10 | 97.55 | 97.18 | 98.52 | 98.66 | 99.13 | 98.68 | 99.12 | 99.12 | 97.66 | 97.73 | 99.10 | 97.90 |
| JSNJ2004-919 | 100 | 97.84 | 98.08 | 94.45 | 93.65 | 95.56 | 95.98 | 97.84 | 98.68 | 98.24 | 97.35 | 95.85 | 96.83 | 98.80 | 97.24 | |
| FJFZ2004-1017 | 100 | 98.14 | 98.32 | 96.95 | 96.97 | 95.56 | 95.98 | 98.70 | 98.68 | 98.09 | 97.79 | 96.08 | 96.60 | 97.03 | 97.81 | |
| JSNT2112-1248 | 99.32 | 97.93 | 98.22 | 96.92 | 96.68 | 95.41 | 95.98 | 98.27 | 98.68 | 97.65 | 97.35 | 96.00 | 96.83 | 98.80 | 97.65 | |
| HNZMD2202-1405 | 99.32 | 98.05 | 98.45 | 97.04 | 96.68 | 95.56 | 95.98 | 97.84 | 97.37 | 97.80 | 97.35 | 95.93 | 96.60 | 99.10 | 97.76 | |
| Recombinant Virus | Parental Virus | Breakpoint 1 | Score for the Seven Detection Methods Embedded in RDP4 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | Region | Begin | End | RDP | GENECONV | BootScan | MaxChi | Chimaera | SiScan | 3Seq | |
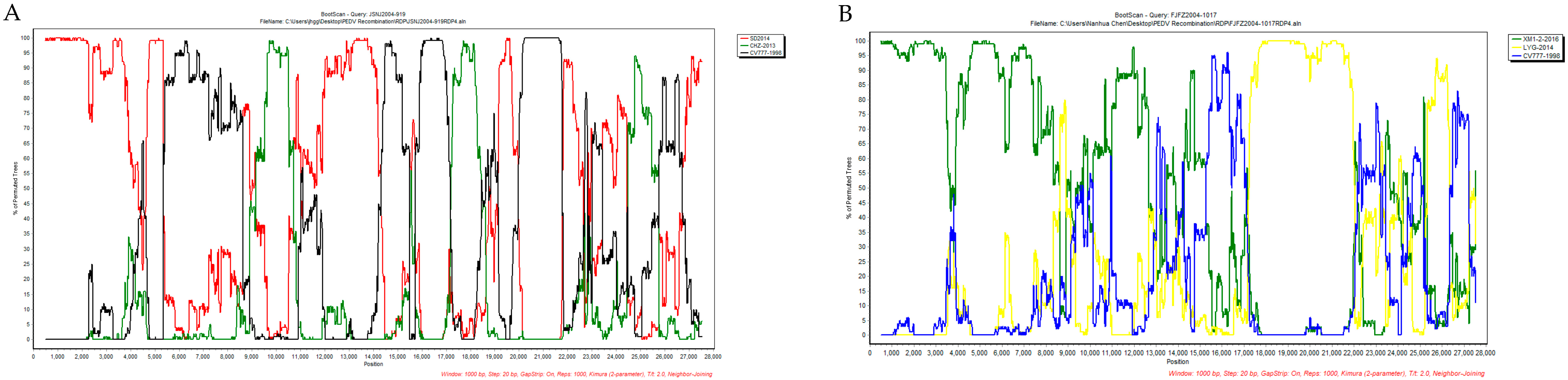
| JSNJ2004-919 | SD2014 (KX064280) | CHZ-2013 (KM609209) | ORF1b | 10,270 | 17,314 | 1.474 × 10−19 | 3.378 × 10−12 | - 2 | 3.691 × 10−7 | 1.611 × 10−7 | 5.710 × 10−11 | 1.509 × 10−13 |
| JSNJ2004-919 | SD2014 (KX064280) | CV777 (KT323979) | ORF1b-S | 15,053 | 21,317 | - | - | - | 3.612 × 10−19 | 3.788 × 10−17 | 5.225 × 10−7 | 8.881 × 10−16 |
| FJFZ2004-1017 | XM1-2 (KX812523) | LYG-2014 (KM609212) | ORF1b-S | 17,651 | 21,558 | 8.796 × 10−29 | 2.824 × 10−32 | 2.835 × 10−34 | 9.863 × 10−20 | 4.821 × 10−15 | 2.963 × 10−24 | 4.440 × 10−16 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, B.; Li, C.; Qiu, Y.; Qi, W.; Qiu, M.; Li, J.; Lin, H.; Zheng, W.; Zhu, J.; Chen, N. Genomic Characterizations of Porcine Epidemic Diarrhea Viruses (PEDV) in Diarrheic Piglets and Clinically Healthy Adult Pigs from 2019 to 2022 in China. Animals 2023, 13, 1562. https://doi.org/10.3390/ani13091562
Feng B, Li C, Qiu Y, Qi W, Qiu M, Li J, Lin H, Zheng W, Zhu J, Chen N. Genomic Characterizations of Porcine Epidemic Diarrhea Viruses (PEDV) in Diarrheic Piglets and Clinically Healthy Adult Pigs from 2019 to 2022 in China. Animals. 2023; 13(9):1562. https://doi.org/10.3390/ani13091562
Chicago/Turabian StyleFeng, Binghui, Chen Li, Yuejia Qiu, Wenhao Qi, Ming Qiu, Jixiang Li, Hong Lin, Wanglong Zheng, Jianzhong Zhu, and Nanhua Chen. 2023. "Genomic Characterizations of Porcine Epidemic Diarrhea Viruses (PEDV) in Diarrheic Piglets and Clinically Healthy Adult Pigs from 2019 to 2022 in China" Animals 13, no. 9: 1562. https://doi.org/10.3390/ani13091562
APA StyleFeng, B., Li, C., Qiu, Y., Qi, W., Qiu, M., Li, J., Lin, H., Zheng, W., Zhu, J., & Chen, N. (2023). Genomic Characterizations of Porcine Epidemic Diarrhea Viruses (PEDV) in Diarrheic Piglets and Clinically Healthy Adult Pigs from 2019 to 2022 in China. Animals, 13(9), 1562. https://doi.org/10.3390/ani13091562








