Canine Mammary Tumor Histopathological Image Classification via Computer-Aided Pathology: An Available Dataset for Imaging Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
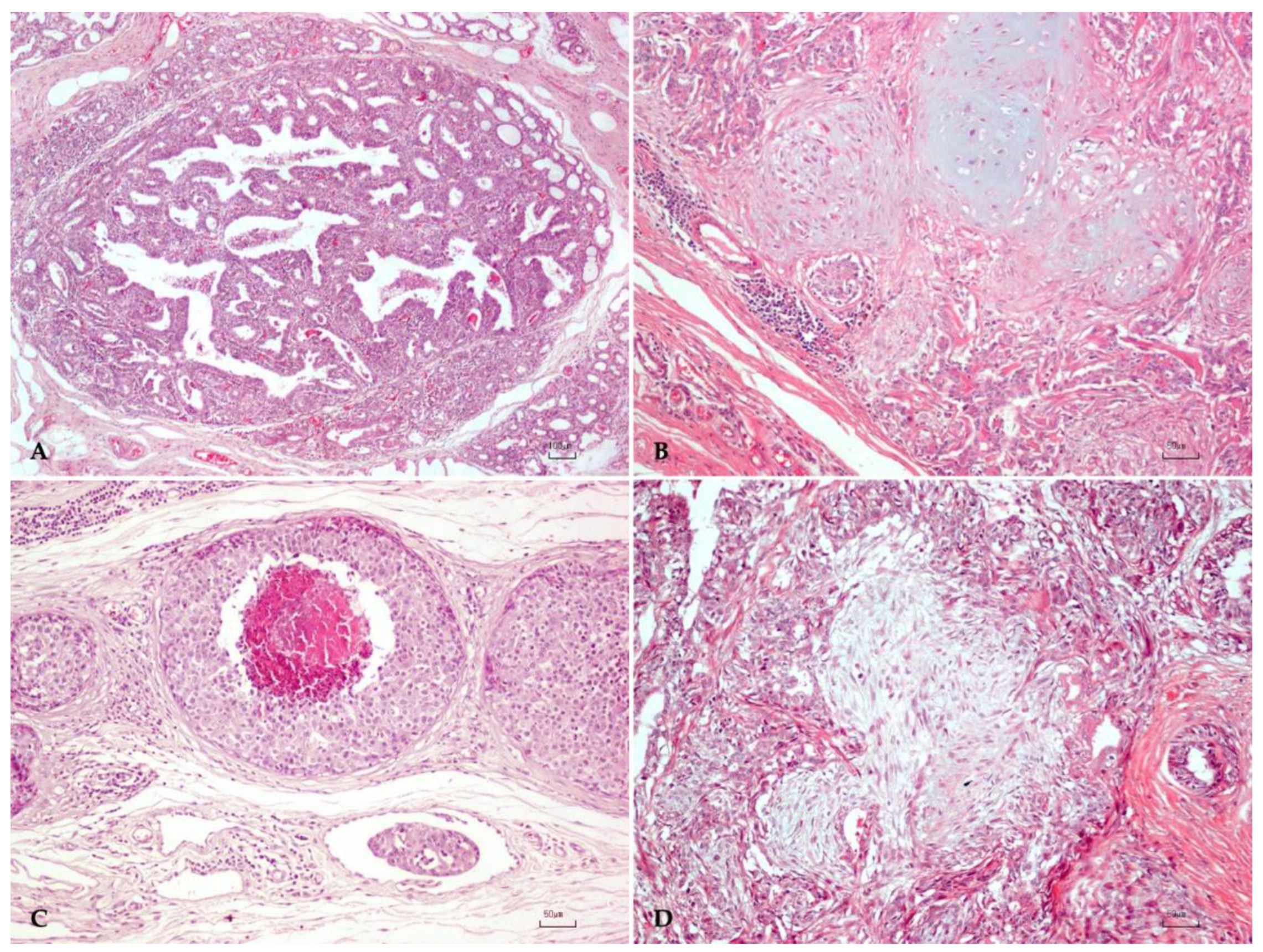
2.1. Canine Mammary Tumor Dataset
2.2. Breast Cancer Dataset
2.3. Data Processing
2.4. Data Augmentation
2.5. Convolutional Neural Networks (CNN)
2.6. Support Vector Machines
2.7. Stochastic Gradient Boosting
3. Results
3.1. Canine Mammary Tumors
3.2. Performance of the Convolutional Neural Networks Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sorenmo, K.U.; Worley, D.R.; Zappulli, V. Tumors of the Mammary Gland. In Withrow & MacEwen’s Small Animal Clinical Oncology, 6th ed.; Withrow, S.J., Vail, D.V., Thamm, D.H., Liptak, J.M., Eds.; Elsevier: St. Louis, MO, USA, 2020; pp. 604–615. [Google Scholar]
- Sleeckx, N.; de Rooster, H.; Veldhuis Kroeze, E.J.; Van Ginneken, C.; Van Brantegem, L. Canine mammary tumours, an overview. Reprod. Domest. Anim. 2011, 46, 1112–1131. [Google Scholar] [CrossRef] [PubMed]
- Papparella, S.; Crescio, M.I.; Baldassarre, V.; Brunetti, B.; Burrai, G.P.; Cocumelli, C.; Grieco, V.; Iussich, S.; Maniscalco, L.; Mariotti, F.; et al. Reproducibility and Feasibility of Classification and National Guidelines for Histological Diagnosis of Canine Mammary Gland Tumours: A Multi-Institutional Ring Study. Vet. Sci. 2022, 9, 357. [Google Scholar] [CrossRef]
- Burrai, G.P.; Gabrieli, A.; Moccia, V.; Zappulli, V.; Porcellato, I.; Brachelente, C.; Pirino, S.; Polinas, M.; Antuofermo, E. A Statistical Analysis of Risk Factors and Biological Behavior in Canine Mammary Tumors: A Multicenter Study. Animals 2020, 10, 1687. [Google Scholar] [CrossRef] [PubMed]
- Antuofermo, E.; Miller, M.A.; Pirino, S.; Xie, J.; Badve, S.; Mohammed, S.I. Spontaneous Mammary Intraepithelial Lesions in Dogs—A Model of Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2007, 16, 2247–2256. [Google Scholar] [CrossRef] [PubMed]
- Mouser, P.; Miller, M.A.; Antuofermo, E.; Badve, S.S.; Mohammed, S.I. Prevalence and Classification of Spontaneous Mammary Intraepithelial Lesions in Dogs without Clinical Mammary Disease. Vet. Pathol. 2010, 47, 275–284. [Google Scholar] [CrossRef]
- Burrai, G.P.; Tanca, A.; De Miglio, M.R.; Abbondio, M.; Pisanu, S.; Polinas, M.; Pirino, S.; Mohammed, S.I.; Uzzau, S.; Addis, M.F.; et al. Investigation of HER2 expression in canine mammary tumors by antibody-based, transcriptomic and mass spectrometry analysis: Is the dog a suitable animal model for human breast cancer? Tumor Biol. 2015, 36, 9083–9091. [Google Scholar] [CrossRef]
- Abdelmegeed, S.M.; Mohammed, S. Canine mammary tumors as a model for human disease. Oncol. Lett. 2018, 15, 8195–8205. [Google Scholar] [CrossRef]
- Mohammed, S.; Meloni, G.; Parpaglia, M.P.; Marras, V.; Burrai, G.; Meloni, F.; Pirino, S.; Antuofermo, E. Mammography and Ultrasound Imaging of Preinvasive and Invasive Canine Spontaneous Mammary Cancer and Their Similarities to Human Breast Cancer. Cancer Prev. Res. 2011, 4, 1790–1798. [Google Scholar] [CrossRef]
- Veta, M.; Pluim, J.P.W.; van Diest, P.J.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Chu, P.Y.; Liao, A.T.; Liu, C.H. Interobserver Variation in the Morphopathological Diagnosis of Canine Mammary Gland Tumor Among Veterinary Pathologists. Int. J. Appl. Res. Vet. Med. 2011, 9, 388–391. [Google Scholar]
- Santos, M.; Correia-Gomes, C.; Santos, A.; de Matos, A.; Dias-Pereira, P.; Lopes, C. Interobserver Reproducibility of Histological Grading of Canine Simple Mammary Carcinomas. J. Comp. Pathol. 2015, 153, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.J.; Brown, R.W.; Bui, M.M.; Chlipala, B.E.A.; Lacchetti, M.C.; Milner, J.D.A.; Pantanowitz, L.; Parwani, A.V.; Reid, M.K.; Riben, M.W.; et al. Validating Whole Slide Imaging Systems for Diagnostic Purposes in Pathology: Guideline Update From the College of American Pathologists in Collaboration With the American Society for Clinical Pathology and the Association for Pathology Informatics. Arch. Pathol. Lab. Med. 2021, 146, 440–450. [Google Scholar] [CrossRef] [PubMed]
- Pallua, J.; Brunner, A.; Zelger, B.; Schirmer, M.; Haybaeck, J. The future of pathology is digital. Pathol. Res. Pract. 2020, 216, 153040. [Google Scholar] [CrossRef] [PubMed]
- Nam, S.; Chong, Y.; Jung, C.K.; Kwak, T.-Y.; Lee, J.Y.; Park, J.; Rho, M.J.; Go, H. Introduction to digital pathology and computer-aided pathology. J. Pathol. Transl. Med. 2020, 54, 125–134. [Google Scholar] [CrossRef]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef]
- Amerikanos, P.; Maglogiannis, I. Image Analysis in Digital Pathology Utilizing Machine Learning and Deep Neural Networks. J. Pers. Med. 2022, 12, 1444. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci. Rep. 2017, 7, srep46450. [Google Scholar] [CrossRef]
- Le Cun, Y.; Jackel, L.; Boser, B.; Denker, J.; Graf, H.; Guyon, I.; Henderson, D.; Howard, R.; Hubbard, W. Handwritten digit recognition: Applications of neural network chips and automatic learning. IEEE Commun. Mag. 1989, 27, 41–46. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y. Deep Learning; MIT Press: Cambridge, MA, USA, 2017; pp. 1–710. [Google Scholar]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012; pp. 1–1049. [Google Scholar]
- Rawat, W.; Wang, Z. Deep Convolutional Neural Networks for Image Classification: A Comprehensive Review. Neural Comput. 2017, 29, 2352–2449. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Correction: Corrigendum: Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 546, 686. [Google Scholar] [CrossRef] [PubMed]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. Breast cancer histopathological image classification using Convolutional Neural Networks. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 2560–2567. [Google Scholar]
- Yang, Y.; Guan, C. Classification of histopathological images of breast cancer using an improved convolutional neural network model. J. X-ray Sci. Technol. 2022, 30, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Spanhol, F.A.; Oliveira, L.S.; Cavalin, P.R.; Petitjean, C.; Heutte, L. Deep features for breast cancer histopathological image classification. In Proceedings of the 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, Canada, 5–8 October 2017; pp. 1868–1873. [Google Scholar] [CrossRef]
- Gupta, V.; Bhavsar, A. Sequential Modeling of Deep Features for Breast Cancer Histopathological Image Classification. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018; pp. 2335–23357. [Google Scholar] [CrossRef]
- Araújo, T.; Aresta, G.; Castro, E.M.; Rouco, J.; Aguiar, P.; Eloy, C.; Polónia, A.; Campilho, A. Classification of breast cancer histology images using Convolutional Neural Networks. PLoS ONE 2017, 12, e0177544. [Google Scholar] [CrossRef] [PubMed]
- Nawaz, M.A.; Sewissy, A.A.; Soliman, T.H.A. Automated Classification of Breast Cancer Histology Images Using Deep Learning Based Convolutional Neural Networks. Int. J. Comput. Sci. Netw. Secur. 2018, 18, 152–160. [Google Scholar]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef]
- Alom, Z.; Yakopcic, C.; Nasrin, M.S.; Taha, T.M.; Asari, V.K. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. J. Digit. Imaging 2019, 32, 605–617. [Google Scholar] [CrossRef]
- Cruz, J.A.; Wishart, D.S. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Inform. 2007, 2, 59–77. [Google Scholar] [CrossRef]
- Yusoff, M.; Haryanto, T.; Suhartanto, H.; Mustafa, W.A.; Zain, J.M.; Kusmardi, K. Accuracy Analysis of Deep Learning Methods in Breast Cancer Classification: A Structured Review. Diagnostics 2023, 13, 683. [Google Scholar] [CrossRef]
- Zappulli, V.; Pena, L.; Rasotto, R.; Goldschmidt, M.H.; Gama, A.; Scruggs, J.L.; Kiupel, M. Volume 2: Mammary Tumors. In Surgical Pathology of Tumors of Domestic Animals; Kiupel, M., Ed.; Davis-Thompson DVM Foundation: Washington, DC, USA, 2019; pp. 1–195. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans. Biomed. Eng. 2016, 63, 1455–1462. [Google Scholar] [CrossRef]
- Breast Cancer Histopathological Database (BreakHis). Available online: https://web.inf.ufpr.br/vri/databases/breast-cancer-histopathological-database-breakhis/ (accessed on 3 May 2021).
- Cawley, G.C.; Talbot, N.L.C. On Over-fitting in Model Selection and Subsequent Selection Bias in Performance Evaluation. J. Mach. Learn. Res. 2010, 11, 2079–2107. [Google Scholar]
- Bussola, N.; Marcolini, A.; Maggio, V.; Jurman, G.; Furlanello, C. Not again! Data Leakage in Digital Pathology. arXiv 2019, arXiv:1909.06539v2. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kumar, A.; Singh, S.K.; Saxena, S.; Lakshmanan, K.; Sangaiah, A.K.; Chauhan, H.; Shrivastava, S.; Singh, R.K. Deep feature learning for histopathological image classification of canine mammary tumors and human breast cancer. Inf. Sci. 2020, 508, 405–421. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A.; Liu, W.; et al. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]
- Tan, M.X.; Le, Q.V. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Volume 97. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Tan, M.; Chen, B.; Pang, R.; Vasudevan, V.; Sandler, M.; Howard, A.; Le, Q.V. MnasNet: Platform-Aware Neural Architecture Search for Mobile. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar] [CrossRef]
- Zheng, L.; Zhao, Y.; Wang, S.; Wang, J.; Tian, Q. Good Practice in CNN Feature Transfer. arXiv 2016, arXiv:1604.00133. [Google Scholar]
- Antropova, N.; Huynh, B.Q.; Giger, M.L. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med. Phys. 2017, 44, 5162–5171. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.Y.H.; Yang, F.; Davis, L.S. Exploiting Local Features from Deep Networks for Image Retrieval. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Liu, Y.; Guo, Y.; Georgiou, T.; Lew, M.S. Fusion that matters: Convolutional fusion networks for visual recognition. Multimedia Tools Appl. 2018, 77, 29407–29434. [Google Scholar] [CrossRef]
- Yang, T.; Li, Y.; Mahdavi, M.; Jin, R.; Zhou, Z. Nystroem Method vs Random Fourier Features: A Theoretical and Empirical Comparison. In Proceedings of the 25th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- Williams, C.K.I.; Seeger, M. Using the Nystroem method to speed up kernel machines. In Proceedings of the 13th International Conference on Neural Information Processing Systems, Denver CO, USA, 1 January 2001. [Google Scholar]
- Pêgo, A.; Aguiar, P. Bioimaging 2015. 2015. Available online: http://www.bioimaging2015.ineb.up.pt/dataset.html (accessed on 5 April 2021).
- Friedman, J.H. Stochastic gradient boosting. Comput. Stat. Data Anal. 2002, 38, 367–378. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer Series in Statistics; Springer: New York, NY, USA, 2009. [Google Scholar]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobot. 2013, 7, 21. [Google Scholar] [CrossRef]
- Friedman, J.H. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Wakili, M.A.; Shehu, H.A.; Sharif, H.; Sharif, H.U.; Umar, A.; Kusetogullari, H.; Ince, I.F.; Uyaver, S. Classification of Breast Cancer Histopathological Images Using DenseNet and Transfer Learning. Comput. Intell. Neurosci. 2022, 2022, 8904768. [Google Scholar] [CrossRef]
- Burrai, G.P.; Baldassarre, V.; Brunetti, B.; Iussich, S.; Maniscalco, L.; Mariotti, F.; Sfacteria, A.; Cocumelli, C.; Grieco, V.; Millanta, F.; et al. Canine and feline in situ mammary carcinoma: A comparative review. Vet. Pathol. 2022, 59, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shen, X.; Zhou, Y.; Wang, X.; Li, T.-Q. Classification of breast cancer histopathological images using interleaved DenseNet with SENet (IDSNet). PLoS ONE 2020, 15, e0232127. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yi, M.; Wu, M.; Wang, J.; He, Y. Breast Pathological Image Classification Based on VGG16 Feature Concatenation. J. Shanghai Jiaotong Univ. 2022, 27, 473–484. [Google Scholar] [CrossRef]
- Dantas Cassali, G.; Cavalheiro Bertagnolli, A.; Ferreira, E.; Araujo Damasceno, K.; de Oliveira Gamba, C.; Bonolo de Campos, C. Canine Mammary Mixed Tumours: A Review. Vet. Med. Int. 2012, 2012, 274608. [Google Scholar] [CrossRef] [PubMed]
- Rajput, D.; Wang, W.-J.; Chen, C.-C. Evaluation of a decided sample size in machine learning applications. BMC Bioinform. 2023, 24, 48. [Google Scholar] [CrossRef] [PubMed]
- Kallipolitis, A.; Revelos, K.; Maglogiannis, I. Ensembling EfficientNets for the Classification and Interpretation of Histopathology Images. Algorithms 2021, 14, 278. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Aubreville, M.; Bertram, C.A.; Marzahl, C.; Gurtner, C.; Dettwiler, M.; Schmidt, A.; Bartenschlager, F.; Merz, S.; Fragoso, M.; Kershaw, O.; et al. Deep learning algorithms out-perform veterinary pathologists in detecting the mitotically most active tumor region. Sci. Rep. 2020, 10, 16447. [Google Scholar] [CrossRef]
- Bertram, C.A.; Klopfleisch, R. The Pathologist 2.0: An Update on Digital Pathology in Veterinary Medicine. Vet. Pathol. 2017, 54, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Rai, T.; Morisi, A.; Bacci, B.; Bacon, N.J.; Dark, M.J.; Aboellail, T.; Thomas, S.A.; Bober, M.; La Ragione, R.; Wells, K. Deep learning for necrosis detection using canine perivascular wall tumour whole slide images. Sci. Rep. 2022, 12, 10634. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Molinari, F.; Iussich, S.; Muscatello, L.V.; Pazzini, L.; Benali, S.; Banco, B.; Abramo, F.; De Maria, R.; Aresu, L. Histopathological Classification of Canine Cutaneous Round Cell Tumors Using Deep Learning: A Multi-Center Study. Front. Vet. Sci. 2021, 8, 640944. [Google Scholar] [CrossRef] [PubMed]
- La Perle, K.M.D. Machine Learning and Veterinary Pathology: Be Not Afraid! Vet. Pathol. 2019, 56, 506–507. [Google Scholar] [CrossRef]
- Awaysheh, A.; Wilcke, J.; Elvinger, F.; Rees, L.; Fan, W.; Zimmerman, K.L. Review of Medical Decision Support and Machine-Learning Methods. Vet. Pathol. 2019, 56, 512–525. [Google Scholar] [CrossRef]
| Feature Extractor | Classifier | Augmentation | Center Crop Accuracy | Ten Crops Accuracy |
|---|---|---|---|---|
| VGG16 | Linear SVM | Advanced 1× | 0.89 ± 0.01 | 0.91 ± 0.01 |
| Inception | Linear SVM | Advanced 6× | 0.88 ± 0.02 | 0.91 ± 0.02 |
| EfficientNet | RBF SVM | Base 6× | 0.9 ± 0.02 | 0.91 ± 0.02 |
| Feature Extractor | Classifier | Augmentation | Center Crop Accuracy | Ten Crops Accuracy |
|---|---|---|---|---|
| VGG16 | Linear SVM | Base 6× | 0.78 ± 0.03 | 0.82 ± 0.03 |
| Inception | Linear SVM | Advanced 6× | 0.78 ± 0.03 | 0.81 ± 0.04 |
| EfficientNet | Poly SVM | Base 6× | 0.84 ± 0.02 | 0.84 ± 0.03 |
| RBF SVM | Base 6× | 0.83 ± 0.03 | 0.85 ± 0.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burrai, G.P.; Gabrieli, A.; Polinas, M.; Murgia, C.; Becchere, M.P.; Demontis, P.; Antuofermo, E. Canine Mammary Tumor Histopathological Image Classification via Computer-Aided Pathology: An Available Dataset for Imaging Analysis. Animals 2023, 13, 1563. https://doi.org/10.3390/ani13091563
Burrai GP, Gabrieli A, Polinas M, Murgia C, Becchere MP, Demontis P, Antuofermo E. Canine Mammary Tumor Histopathological Image Classification via Computer-Aided Pathology: An Available Dataset for Imaging Analysis. Animals. 2023; 13(9):1563. https://doi.org/10.3390/ani13091563
Chicago/Turabian StyleBurrai, Giovanni P., Andrea Gabrieli, Marta Polinas, Claudio Murgia, Maria Paola Becchere, Pierfranco Demontis, and Elisabetta Antuofermo. 2023. "Canine Mammary Tumor Histopathological Image Classification via Computer-Aided Pathology: An Available Dataset for Imaging Analysis" Animals 13, no. 9: 1563. https://doi.org/10.3390/ani13091563
APA StyleBurrai, G. P., Gabrieli, A., Polinas, M., Murgia, C., Becchere, M. P., Demontis, P., & Antuofermo, E. (2023). Canine Mammary Tumor Histopathological Image Classification via Computer-Aided Pathology: An Available Dataset for Imaging Analysis. Animals, 13(9), 1563. https://doi.org/10.3390/ani13091563





