Molecular Detection and Genetic Characterization of Vertically Transmitted Viruses in Ducks
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. Viral DNA/RNA Extraction and Reverse Transcription
2.3. PCR
2.4. DNA Sequences and Phylogenetic Analysis
3. Results
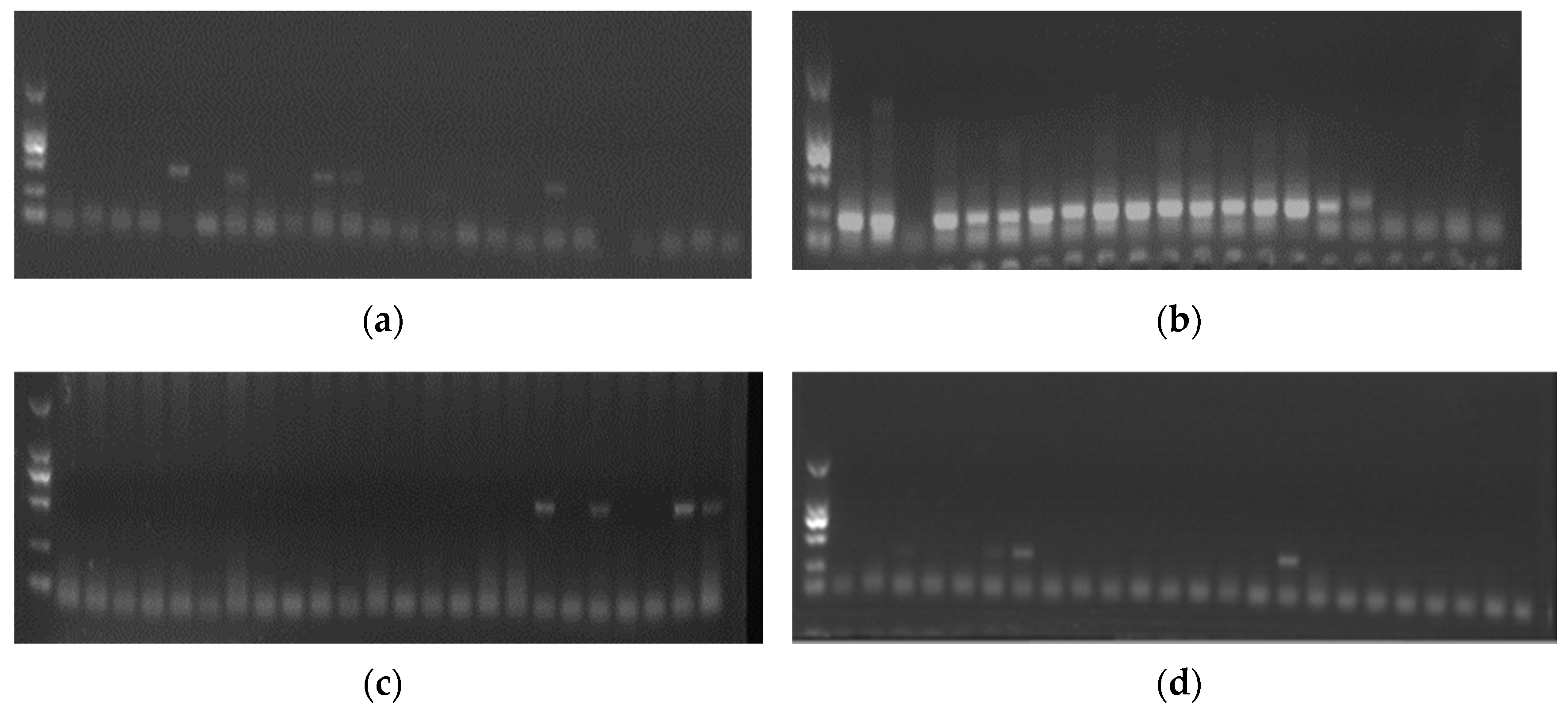
3.1. Prevalence of DHBV, DuCV, DHAV-3, and ARV in Ducks
3.2. Coinfection of DHBV, DuCV, DHAV-3, and ARV
3.3. Sequencing and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Patil, S.S.; Shinduja, R.; Suresh, K.P.; Phukan, S.; Kumar, S.; Sengupta, P.P.; Amachawadi, R.G.; Raut, A.; Roy, P.; Syed, A.; et al. A systematic review and meta-analysis on the prevalence of infectious diseases of Duck: A world perspective. Saudi J. Biol. Sci. 2021, 28, 5131–5144. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, X.; Zhang, R.; Chen, J.; Xia, L.; Lin, S.; Xie, Z.; Jiang, S. Evidence of possible vertical transmission of duck circovirus. Vet. Microbiol. 2014, 174, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, M.; Robinson, W.S.; Marion, P.L. Duck hepatitis B virus replicates in the yolk sac of developing embryos. J. Virol. 1987, 61, 2273–2279. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Y.; Gao, B.; Zhang, S.; Diao, Y.; Tang, Y. Evidence of vertical transmission of novel duck orthoreovirus in ducks. Vet. Microbiol. 2020, 251, 108861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Yang, Y.; Lan, J.; Xie, Z.; Zhang, X.; Jiang, S. Evidence of possible vertical transmission of duck hepatitis A virus type 1 in ducks. Transbound. Emerg. Dis. 2021, 68, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, S.; Liu, W.; Hu, Z. Current status and future direction of duck hepatitis A virus vaccines. Avian Pathol. 2023, 52, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Mason, W.S.; Seal, G.; Summers, J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J. Virol. 1980, 36, 829–836. [Google Scholar] [CrossRef] [PubMed]
- Uchida, T.; Suzuki, K.; Esumi, M.; Arii, M.; Shikata, T. Influence of aflatoxin B1 intoxication on duck livers with duck hepatitis B virus infection. Cancer Res. 1988, 48, 1559–1565. [Google Scholar]
- Zhang, D.; Wang, Y.; He, Y.; Ji, L.; Zhao, K.; Yang, S.; Zhang, W. Identification of avihepadnaviruses and circoviruses in an unexplained death event in farmed ducks. Arch. Virol. 2023, 168, 85. [Google Scholar] [CrossRef]
- Liu, Q.; Jia, R.; Wang, M.; Huang, J.; Zhu, D.; Chen, S.; Yin, Z.; Wang, Y.; Chen, X.; Cheng, A. Cloning, expression and purification of duck hepatitis B virus (DHBV) core protein and its use in the development of an indirect ELISA for serologic detection of DHBV infection. Arch. Virol. 2014, 159, 897–904. [Google Scholar] [CrossRef]
- Tan, Y.; Liu, J.; Qin, Y.; Liang, B.; Gu, Y.; Liang, L.; Liu, L.; Liu, Y.; Su, H. Glucose Homeostasis Is Dysregulated in Ducks Infected with Duck Hepatitis B Virus. Intervirology 2021, 64, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Zhou, D.; Liu, J.; Hao, X.; Cheng, Z. Duck circovirus induces a new pathogenetic characteristic, primary sclerosing cholangitis. Comp. Immunol. Microbiol. Infect. Dis. 2019, 63, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Liu, N.; Zhang, L.; Jiang, W.; Fan, X.; Wang, X.; Miao, R.; Zhai, X.; Wei, L.; Jiang, S.; et al. Research Note: Complete genome cloning and genetic evolution analysis of four Cherry Valley duck circovirus strains in China in 2022. Poult. Sci. 2023, 102, 102920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jia, R.; Lu, Y.; Wang, M.; Zhu, D.; Chen, S.; Yin, Z.; Chen, X.; Cheng, A. Identification, genotyping, and molecular evolution analysis of duck circovirus. Gene 2013, 529, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Wu, Y.; Yang, C.; Zhang, X.; Lian, C.; Chen, H.; Han, L. Comments on duck circovirus (DuCV) genotype definition. Gene 2014, 538, 207–208. [Google Scholar] [CrossRef]
- Shen, M.; Gao, P.; Wang, C.; Li, N.; Zhang, S.; Jiang, Y.; Liu, D.; Jia, B.; Xu, L.; Huang, B.; et al. Pathogenicity of duck circovirus and fowl adenovirus serotype 4 co-infection in Cherry Valley ducks. Vet. Microbiol. 2023, 279, 109662. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, L.X.; Sun, W.C.; Shi, N.; Sun, X.T.; Jin, N.Y.; Si, X.K. Molecular survey of duck circovirus infection in poultry in southern and southwestern China during 2018 and 2019. BMC Vet. Res. 2020, 16, 80. [Google Scholar] [CrossRef]
- Li, P.; Li, J.; Zhang, R.; Chen, J.; Wang, W.; Lan, J.; Xie, Z.; Jiang, S. Duck “beak atrophy and dwarfism syndrome” disease complex: Interplay of novel goose parvovirus-related virus and duck circovirus? Transbound. Emerg. Dis. 2018, 65, 345–351. [Google Scholar] [CrossRef]
- Banda, A.; Galloway-Haskins, R.I.; Sandhu, T.S.; Schat, K.A. Genetic analysis of a duck circovirus detected in commercial Pekin ducks in New York. Avian Dis. 2007, 51, 90–95. [Google Scholar] [CrossRef]
- Chen, C.L.; Wang, P.X.; Lee, M.S.; Shien, J.H.; Shien, H.K.; Ou, S.J.; Chen, C.H.; Chang, P.C. Development of a polymerase chain reaction procedure for detection and differentiation of duck and goose circovirus. Avian Dis. 2006, 50, 92–95. [Google Scholar] [CrossRef]
- Matczuk, A.K.; Krawiec, M.; Wieliczko, A. A new duck circovirus sequence, detected in velvet scoter (Melanitta fusca) supports great diversity among this species of virus. Virol. J. 2015, 12, 121. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.Y.; Xiong, W.J.; Tang, H.; Xiao, C.T. Identification and characterization of a novel circovirus species in domestic laying ducks designated as duck circovirus 3 (DuCV3) from Hunan province, China. Vet. Microbiol. 2022, 275, 109598. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, Y.; Cheng, A.; Wang, M.; Liu, M.; Zhu, D.; Chen, S.; Zhao, X.; Yang, Q.; Wu, Y.; et al. Duck Circovirus genotype 2 ORF3 protein induces apoptosis through the mitochondrial pathway. Poult. Sci. 2023, 102, 102533. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Cheng, A.; Wang, M.; Ou, X.; Sun, D.; Zhang, S.; Mao, S.; Yang, Q.; Tian, B.; Wu, Y.; et al. DHAV 3CD targets IRF7 and RIG-I proteins to block the type I interferon upstream signaling pathway. Vet. Res. 2023, 54, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, M.; Zhou, S.; Cheng, A.; Ou, X.; Sun, D.; Wu, Y.; Yang, Q.; Gao, Q.; Huang, J.; et al. The DHAV-1 protein VP1 interacts with PI3KC3 to induce autophagy through the PI3KC3 complex. Vet. Res. 2022, 53, 64. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Cheng, A.; Wang, M.; Jia, R.; Zhu, D.; Chen, S.; Liu, M.; Sun, K.; Yang, Q.; Wu, Y.; et al. Recent advances from studies on the role of structural proteins in enterovirus infection. Future Microbiol. 2015, 10, 1529–1542. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Ou, X.; Zhu, D.; Ma, G.; Cheng, A.; Wang, M.; Chen, S.; Jia, R.; Liu, M.; Sun, K.; et al. The 2A2 protein of Duck hepatitis A virus type 1 induces apoptosis in primary cell culture. Virus Genes 2016, 52, 780–788. [Google Scholar] [CrossRef]
- Sun, D.; Chen, S.; Cheng, A.; Wang, M. Roles of the Picornaviral 3C Proteinase in the Viral Life Cycle and Host Cells. Viruses 2016, 8, 82. [Google Scholar] [CrossRef]
- Sun, D.; Wang, M.; Wen, X.; Cheng, A.; Jia, R.; Sun, K.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; et al. Cleavage of poly(A)-binding protein by duck hepatitis A virus 3C protease. Sci. Rep. 2017, 7, 16261. [Google Scholar] [CrossRef]
- Yang, C.; Shah, P.T.; Bahoussi, A.N.; Wu, C.; Wang, L.; Xing, L. Duck hepatitis a virus: Full-length genome-based phylogenetic and phylogeographic view during 1986–2020. Virus Res. 2023, 336, 199216. [Google Scholar] [CrossRef]
- Zhou, S.; Li, S.; Wang, Y.; Li, X.; Zhang, T. Duck hepatitis A virus prevalence in mainland China between 2009 and 2021: A systematic review and meta-analysis. Prev. Vet. Med. 2022, 208, 105730. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.H.; Tsai, H.J. Molecular characterization of a new serotype of duck hepatitis virus. Virus Res. 2007, 126, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Kwon, Y.K.; Joh, S.J.; Kim, S.J.; Tolf, C.; Kim, J.H.; Sung, H.W.; Lindberg, A.M.; Kwon, J.H. Recent Korean isolates of duck hepatitis virus reveal the presence of a new geno- and serotype when compared to duck hepatitis virus type 1 type strains. Arch. Virol. 2007, 152, 2059–2072. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.J.; Cheng, A.C.; Wang, M.S.; Jia, R.Y.; Zhu, D.K.; Chen, S.; Liu, M.F.; Liu, F.; Chen, X.Y. Detection, differentiation, and VP1 sequencing of duck hepatitis A virus type 1 and type 3 by a 1-step duplex reverse-transcription PCR assay. Poult. Sci. 2014, 93, 2184–2192. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Zhu, D.; Cheng, A.; Wang, M.; Chen, S.; Jia, R.; Liu, M.; Sun, K.; Zhao, X.; Yang, Q.; et al. Molecular epidemiology of duck hepatitis a virus types 1 and 3 in China, 2010–2015. Transbound. Emerg. Dis. 2018, 65, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Xia, X.; Cheng, A.; Wang, M.; Ou, X.; Mao, S.; Sun, D.; Zhang, S.; Yang, Q.; Wu, Y.; et al. DHAV-1 Blocks the Signaling Pathway Upstream of Type I Interferon by Inhibiting the Interferon Regulatory Factor 7 Protein. Front. Microbiol. 2021, 12, 700434. [Google Scholar] [CrossRef]
- Wu, F.; Lu, F.; Fan, X.; Chao, J.; Liu, C.; Pan, Q.; Sun, H.; Zhang, X. Immune-related miRNA-mRNA regulation network in the livers of DHAV-3-infected ducklings. BMC Genom. 2020, 21, 123. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Y.; Chen, Y.; Liang, S.; Liu, D.; Fan, W.; Xu, Y.; Liu, H.; Zhou, Z.; Liu, X.; et al. Dynamic Transcriptome Reveals the Mechanism of Liver Injury Caused by DHAV-3 Infection in Pekin Duck. Front. Immunol. 2020, 11, 568565. [Google Scholar] [CrossRef]
- Jones, R.C. Avian reovirus infections. Rev. Sci. Tech. 2000, 19, 614–625. [Google Scholar] [CrossRef]
- Varela, R.; Benavente, J. Protein coding assignment of avian reovirus strain S1133. J. Virol. 1994, 68, 6775–6777. [Google Scholar] [CrossRef]
- Farnoushi, Y.; Heller, D.; Lublin, A. Development of a wide-range real-time RT-PCR assay for detection of Avian reovirus (ARV). J. Virol. Methods 2022, 310, 114613. [Google Scholar] [CrossRef] [PubMed]
- Benavente, J.; Martínez-Costas, J. Avian reovirus: Structure and biology. Virus Res. 2007, 123, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zou, Z.; Song, S.; Liu, H.; Gong, X.; Li, B.; Liu, P.; Wang, Q.; Liu, F.; Luan, D.; et al. Epidemiological Analysis of Avian Reovirus in China and Research on the Immune Protection of Different Genotype Strains from 2019 to 2020. Vaccines 2023, 11, 485. [Google Scholar] [CrossRef] [PubMed]
- Egana-Labrin, S.; Broadbent, A.J. Avian reovirus: A furious and fast evolving pathogen. J. Med. Microbiol. 2023, 72, 001761. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, D. Avian Reovirus in Israel, Variants and Vaccines-A Review. Avian Dis. 2022, 66, 447–451. [Google Scholar] [CrossRef]
- Goldenberg, D.; Pasmanik-Chor, M.; Pirak, M.; Kass, N.; Lublin, A.; Yeheskel, A.; Heller, D.; Pitcovski, J. Genetic and antigenic characterization of sigma C protein from avian reovirus. Avian Pathol. 2010, 39, 189–199. [Google Scholar] [CrossRef]
- Kant, A.; Balk, F.; Born, L.; van Roozelaar, D.; Heijmans, J.; Gielkens, A.; ter Huurne, A. Classification of Dutch and German avian reoviruses by sequencing the sigma C protein. Vet. Res. 2003, 34, 203–212. [Google Scholar] [CrossRef]
- Liu, H.J.; Lee, L.H.; Shih, W.L.; Li, Y.J.; Su, H.Y. Rapid characterization of avian reoviruses using phylogenetic analysis, reverse transcription-polymerase chain reaction and restriction enzyme fragment length polymorphism. Avian Pathol. 2004, 33, 171–180. [Google Scholar] [CrossRef]
- Troxler, S.; Rigomier, P.; Bilic, I.; Liebhart, D.; Prokofieva, I.; Robineau, B.; Hess, M. Identification of a new reovirus causing substantial losses in broiler production in France, despite routine vaccination of breeders. Vet. Rec. 2013, 172, 556. [Google Scholar] [CrossRef]
- Lu, H.; Tang, Y.; Dunn, P.A.; Wallner-Pendleton, E.A.; Lin, L.; Knoll, E.A. Isolation and molecular characterization of newly emerging avian reovirus variants and novel strains in Pennsylvania, USA, 2011–2014. Sci. Rep. 2015, 5, 14727. [Google Scholar] [CrossRef]
- Ayalew, L.E.; Gupta, A.; Fricke, J.; Ahmed, K.A.; Popowich, S.; Lockerbie, B.; Tikoo, S.K.; Ojkic, D.; Gomis, S. Phenotypic, genotypic and antigenic characterization of emerging avian reoviruses isolated from clinical cases of arthritis in broilers in Saskatchewan, Canada. Sci. Rep. 2017, 7, 3565. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Shang, H.; Zhou, F.; Wang, C.; Zhang, S.; Gao, P.; Guo, P.; Zhu, R.; Sun, Z.; et al. Effects of duck circovirus on immune function and secondary infection of Avian Pathogenic Escherichia coli. Poult. Sci. 2022, 101, 101799. [Google Scholar] [CrossRef]
- Hattermann, K.; Schmitt, C.; Soike, D.; Mankertz, A. Cloning and sequencing of Duck circovirus (DuCV). Arch. Virol. 2003, 148, 2471–2480. [Google Scholar] [CrossRef] [PubMed]
- Fringuelli, E.; Scott, A.N.; Beckett, A.; McKillen, J.; Smyth, J.A.; Palya, V.; Glavits, R.; Ivanics, E.; Mankertz, A.; Franciosini, M.P.; et al. Diagnosis of duck circovirus infections by conventional and real-time polymerase chain reaction tests. Avian Pathol. 2005, 34, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.Y.; Kang, M.; Cho, J.G.; Jang, H.K. Genetic analysis of duck circovirus in Pekin ducks from South Korea. Poult. Sci. 2013, 92, 2886–2891. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xie, X.; Zhang, D.; Ma, G.; Wang, X.; Zhang, D. Detection of duck circovirus in China: A proposal on genotype classification. Vet. Microbiol. 2011, 147, 410–415. [Google Scholar] [CrossRef]
- Soike, D.; Albrecht, K.; Hattermann, K.; Schmitt, C.; Mankertz, A. Novel circovirus in mulard ducks with developmental and feathering disorders. Vet. Rec. 2004, 154, 792–793. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, W.; Zhang, F.; Li, Y.; Li, J.; Liang, S.; Yu, X.; Peng, C.; Liu, S.; Wang, J.; et al. Development of a dual-labeled, hydrolysis probe-based, real-time quantitative PCR assay for detection of both genotypes of duck circovirus-1 (DuCV-1) and DuCV-2. Virus Genes 2021, 57, 453–458. [Google Scholar] [CrossRef]
- Wan, C.H.; Fu, G.H.; Shi, S.H.; Cheng, L.F.; Chen, H.M.; Peng, C.X.; Lin, S.; Huang, Y. Epidemiological investigation and genome analysis of duck circovirus in Southern China. Virol. Sin. 2011, 26, 289–296. [Google Scholar] [CrossRef]
- Hong, Y.T.; Kang, M.; Jang, H.K. Pathogenesis of duck circovirus genotype 1 in experimentally infected Pekin ducks. Poult. Sci. 2018, 97, 3050–3057. [Google Scholar] [CrossRef]
- Liu, S.N.; Zhang, X.X.; Zou, J.F.; Xie, Z.J.; Zhu, Y.L.; Zhao, Q.; Zhou, E.M.; Jiang, S.J. Development of an indirect ELISA for the detection of duck circovirus infection in duck flocks. Vet. Microbiol. 2010, 145, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, S.; Wu, J.; Zhao, Q.; Sun, Y.; Kong, Y.; Li, X.; Yao, M.; Chai, T. An investigation of duck circovirus and co-infection in Cherry Valley ducks in Shandong Province, China. Vet. Microbiol. 2009, 133, 252–256. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Liu, L.; Han, C.; Li, J.; Zeng, X. First findings of duck circovirus in migrating wild ducks in China. Vet. Microbiol. 2018, 216, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zheng, M.; Cao, H.; Lu, H.; Wei, X.; Pan, Y.; Zhang, H.; Su, J.; Li, J.; Jin, N. Genome Sequences of a Novel Recombinant Duck Circovirus in China. Genome Announc. 2016, 4, e01181-16. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, G.L.; Vidigal, P.M.P.; Fietto, J.L.R.; Bressan, G.C.; Silva Júnior, A.; de Almeida, M.R. Evolutionary analysis of Porcine circovirus 3 (PCV3) indicates an ancient origin for its current strains and a worldwide dispersion. Virus Genes 2018, 54, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Stenzel, T.; Dziewulska, D.; Muhire, B.M.; Hartnady, P.; Kraberger, S.; Martin, D.P.; Varsani, A. Recombinant Goose Circoviruses Circulating in Domesticated and Wild Geese in Poland. Viruses 2018, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Lin, Z.; Dai, A.; Chen, H.; Ma, Y.; Li, N.; Wu, Y.; Yang, X.; Luo, M.; Liu, J. Emergence of a novel recombinant porcine circovirus type 2 in China: PCV2c and PCV2d recombinant. Transbound. Emerg. Dis. 2019, 66, 2496–2506. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D.; Bai, C.X.; Guo, X.; Gao, W.H.; Li, M.L.; Wang, J.; Li, Y.D. Molecular characteristics of a novel duck circovirus subtype 1d emerging in Anhui, China. Virus Res. 2021, 295, 198216. [Google Scholar] [CrossRef]
- Li, Z.; Fu, G.; Feng, Z.; Chen, J.; Shi, S.; Liu, R.; Cheng, L.; Chen, H.; Wan, C.; Yu, H. Evaluation of a novel inactivated vaccine against duck circovirus in muscovy ducks. Vet. Microbiol. 2020, 241, 108574. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, S.; Shang, H.; Wang, C.; Zhou, F.; Liu, Y.; Jiang, Y.; Gao, P.; Li, N.; Liu, D.; et al. Evaluation of the antiviral effect of four plant polysaccharides against duck circovirus. Res. Vet. Sci. 2022, 152, 446–457. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, C.; Qu, Z.; Zhang, W.; Liu, Y.; Qi, H.; Hao, C.; Zhang, W.; Gao, M.; Wang, J.; et al. Pathogenicity of duck hepatitis A virus type 3 and innate immune responses of the ducklings to virulent DHAV-3. Mol. Immunol. 2018, 95, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Saikia, D.P.; Yadav, K.; Pathak, D.C.; Ramamurthy, N.; D’Silva, A.L.; Marriappan, A.K.; Ramakrishnan, S.; Vakharia, V.N.; Chellappa, M.M.; Dey, S. Recombinant Newcastle Disease Virus (NDV) Expressing Sigma C Protein of Avian Reovirus (ARV) Protects against Both ARV and NDV in Chickens. Pathogens 2019, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- Shih, W.L.; Hsu, H.W.; Liao, M.H.; Lee, L.H.; Liu, H.J. Avian reovirus sigmaC protein induces apoptosis in cultured cells. Virology 2004, 321, 65–74. [Google Scholar] [CrossRef]
- Fehér, E.; Jakab, S.; Bali, K.; Kaszab, E.; Nagy, B.; Ihász, K.; Bálint, Á.; Palya, V.; Bányai, K. Genomic Epidemiology and Evolution of Duck Hepatitis A Virus. Viruses 2021, 13, 1592. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.M.G.; Mohamed, F.F.; ElBakrey, R.M.; Eid, A.A.M.; Mor, S.K.; Goyal, S.M. Outbreaks of Duck Hepatitis A Virus in Egyptian Duckling Flocks. Avian Dis. 2019, 63, 68–74. [Google Scholar] [CrossRef]
- Liu, H.J.; Lee, L.H.; Hsu, H.W.; Kuo, L.C.; Liao, M.H. Molecular evolution of avian reovirus: Evidence for genetic diversity and reassortment of the S-class genome segments and multiple cocirculating lineages. Virology 2003, 314, 336–349. [Google Scholar] [CrossRef]
- Cavanagh, D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007, 38, 281–297. [Google Scholar] [CrossRef]
- Egaña-Labrin, S.; Hauck, R.; Figueroa, A.; Stoute, S.; Shivaprasad, H.L.; Crispo, M.; Corsiglia, C.; Zhou, H.; Kern, C.; Crossley, B.; et al. Genotypic Characterization of Emerging Avian Reovirus Genetic Variants in California. Sci. Rep. 2019, 9, 9351. [Google Scholar] [CrossRef]




| Virus | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Fragment Size (bp) |
|---|---|---|---|
| DuCV | gcacgctcgacaattgcaagt | gccacgcccaaagattacataag | 338 |
| DHBV | gggctaggagattgctttg | ggttcgagtccacgaggtt | 217 |
| DHAV-1 | agcttaaggcccggtgccccg | ggtagggtagggaatagtaaagt | 399 |
| DHAV-3 | aacccctttgatccacactg | gataaggcatccacaccatc | 544 |
| ARV | taatttagacggtttgagga | cgttgagaacagaagtaggg | 324 |
| Virus | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Fragment Size (bp) |
|---|---|---|---|
| DuCV | accggcgcttgtactccgtactcc | aataatataacggcgcttgtgcggt | 1995 |
| DHAV-3-VP1 | ggtgattccaatcagcttggcga | ttcaatttctagatggagctcaaag | 720 |
| ARV-σC | atggcgggtctcaatccatcgca | ttaggtgtcgatgccggtacgcacg | 981 |
| Duck Breed | Number of Samples | Positive Sample (%) | |||
|---|---|---|---|---|---|
| DHBV | DuCV | DHAV-3 | ARV | ||
| Jinding | 152 | 63.15 (96/152) | 53.95 (82/152) | 8.55 (13/152) | 18.42 (28/152) |
| Shaoxing | 119 | 78.15 (93/119) | 21.01 (25/119) | 8.40 (10/119) | 21.85 (26/119) |
| Overall | 271 | 69.74 (189/271) | 39.48 (107/271) | 8.49 (23/271) | 19.92 (54/271) |
| Virus | Coinfection Rate (%) | Virus | Coinfection Rate (%) |
|---|---|---|---|
| DHBV and DuCV | 22.75 | DHBV and ARV | 10.20 |
| DHBV and DHAV-3 | 5.49 | DHBV, DHAV-3 and ARV | 0.78 |
| DuCV and ARV | 10.98 | DHBV, DuCV and DHAV-3 | 1.96 |
| DuCV and DHAV-3 | 3.92 | DuCV, DHAV-3 and ARV | 0.78 |
| DHAV-3 and ARV | 1.96 | DHBV, DuCV, DHAV-3 and ARV | 0.78 |
| No | Strain | Year | Region | Accession No. | Cap Gene Sequence Identity (%) | Rep Gene Sequence Identity (%) | Full Genome Identity (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | |||||
| 1 | DU095 | 2010 | Beijing/CN | HM162349.1 | 94.1% | 96.5% | 98.2% | 97.6% | 96.2% |
| 2 | TAYB01 | 2018 | Shandong/CN | MH444647.1 | 93.9% | 96.5% | 98.4% | 96.9% | 96.4% |
| 3 | D12-KD-001 | 2013 | Korea | KC851817.1 | 94.4% | 96.9% | 98.2% | 98.0% | 96.4% |
| 4 | AH01 | 2020 | Henan/CN | MN928808.1 | 94.4% | 96.5% | 99.1% | 97.6% | 97.3% |
| 5 | SDFC12 | 2016 | Shandong/CN | KY328304.1 | 94.2% | 96.5% | 98.5% | 97.3% | 97.1% |
| 6 | VC4 | 2020 | Brazil | MT318126.1 | 93.5% | 96.5% | 98.2% | 97.3% | 96.4% |
| 7 | - | 2005 | USA | NC007220.1 | 93.5% | 96.5% | 98.1% | 96.9% | 96.2% |
| 8 | 33753-52 | 2005 | USA | DQ100076.1 | 93.5% | 96.5% | 98.1% | 96.9% | 96.2% |
| 9 | D11-JW-008 | 2012 | Korea | JQ740363.1 | 94.7% | 96.9% | 98.2% | 97.6% | 97.0% |
| 10 | HN02 | 2020 | Henan/CN | MN928795.1 | 94.4% | 96.5% | 99.2% | 99.0% | 97.4% |
| 11 | NN11/2012 | 2013 | Guangxi/CN | KC460530.1 | 96.5% | 96.1% | 99.0% | 99.0% | 98.0% |
| 12 | DB41-18 | 2014 | Taiwan/CN | KP229368.1 | 96.4% | 97.3% | 98.3% | 98.0% | 97.4% |
| 13 | WF0803 | 2009 | Shandong/CN | GU131342.1 | 97.2% | 97.7% | 99.2% | 99.0% | 97.8% |
| 14 | WF0701 | 2007 | Shandong/CN | EU022375.1 | 96.9% | 97.3% | 98.9% | 99.0% | 97.6% |
| 15 | ShijiazhuangNo. 3 | 2020 | Hebei/CN | MW255979.1 | 97.2% | 97.3% | 98.8% | 98.6% | 97.9% |
| 16 | LA-18 | 2020 | Hebei/CN | MT799793.1 | 96.6% | 97.3% | 98.5% | 98.6% | 97.6% |
| 17 | D12-MR-020 | 2013 | Korea | KC851822.1 | 97.3% | 98.8% | 98.8% | 99.0% | 98.0% |
| 18 | SDHZ1223 | 2020 | Shandong/CN | MT084134.1 | 99.9% | 100.0% | 99.9% | 99.7% | 99.7% |
| 19 | BaodingNO.Du | 2020 | Hebei/CN | MW255980.1 | 99.7% | 100.0% | 99.7% | 99.7% | 99.7% |
| 20 | HZ170301 | 2019 | Shandong/CN | MN068356.1 | 99.9% | 100.0% | 99.8% | 99.7% | 99.8% |
| 21 | QD | 2021 | QingDao/CN | MZ425925.1 | 99.6% | 99.6% | 99.3% | 98.6% | 99.5% |
| 22 | WF190501 | 2019 | Shandong/CN | MN068359.1 | 99.9% | 100.0% | 99.3% | 99.0% | 99.4% |
| 23 | GD/ZQ/144 | 2022 | Guangdong/CN | ON227548.1 | 100.0% | 100.0% | 99.9% | 99.7% | 99.8% |
| 24 | GD/QY/145 | 2022 | Guangdong/CN | ON227549.1 | 100.0% | 100.0% | 99.5% | 99.3% | 99.7% |
| 25 | GD/FS/125 | 2022 | Guangdong/CN | ON227538.1 | 99.6% | 99.6% | 99.2% | 98.3% | 99.4% |
| 26 | GD/FS/132 | 2022 | Guangdong/CN | ON227544.1 | 99.5% | 98.8% | 99.5% | 98.6% | 99.6% |
| 27 | GD/JM/127 | 2022 | Guangdong/CN | ON227540.1 | 99.4% | 100.0% | 97.0% | 95.9% | 97.6% |
| 28 | JSPX03B | 2017 | Shandong/CN | MF627687.1 | 92.2% | 97.3% | 97.6% | 97.6% | 95.5% |
| 29 | QZ36/2012 | 2013 | Guangxi/CN | KC460534.1 | 91.0% | 97.7% | 96.7% | 95.9% | 94.3% |
| 30 | SDDY0520 | 2017 | Shandong/CN | MF627689.1 | 93.2% | 96.9% | 96.8% | 96.9% | 95.1% |
| 31 | FJZZ302 | 2009 | Fujian/CN | GQ423747.1 | 92.2% | 96.9% | 97.8% | 98.0% | 95.2% |
| 32 | CD13056 | 2014 | Taiwan/CN | KP229375.1 | 90.2% | 96.5% | 97.4% | 98.0% | 94.3% |
| 33 | DB7-17 | 2014 | Taiwan/CN | KP229364.1 | 89.5% | 95.7% | 97.4% | 98.0% | 94.0% |
| 34 | MH02/07 | 2008 | Fujian/CN | EU499309.1 | 79.1% | 89.1% | 92.4% | 85.7% | 84.8% |
| 35 | HZ09 | 2007 | Fujian/CN | EU344802.1 | 78.9% | 89.1% | 92.3% | 85.7% | 85.2% |
| 36 | GD190403 | 2019 | Guangdong/CN | MK814577.1 | 78.9% | 88.4% | 88.2% | 82.9% | 82.9% |
| 37 | 31-1 | 2013 | Guangdong/CN | KF941312.1 | 79.1% | 88.4% | 88.2% | 82.9% | 82.9% |
| 38 | LY0701 | 2007 | Shandong/CN | EU022374.1 | 78.7% | 87.6% | 87.8% | 81.2% | 82.7% |
| 39 | CD0912 | 2021 | Chengdu/CN | OK094646.1 | 79.1% | 88.4% | 87.8% | 83.3% | 82.8% |
| 40 | YB02 | 2021 | Chengdu/CN | OK094645.1 | 79.1% | 88.4% | 87.5% | 82.3% | 82.7% |
| 41 | FJ1815 | 2019 | Fujian/CN | MN052853.1 | 78.9% | 88.0% | 87.5% | 81.6% | 82.7% |
| 42 | YN180506 | 2019 | Guangdong/CN | MK814585.1 | 78.4% | 87.2% | 87.6% | 82.3% | 82.4% |
| 43 | PT07 | 2008 | Fujian/CN | EU499310.1 | 79.1% | 88.4% | 87.5% | 80.9% | 82.8% |
| 44 | AHAU28 | 2020 | Anhui/CN | MT646348.1 | 78.8% | 88.0% | 87.6% | 81.6% | 82.7% |
| 45 | GX1104 | 2012 | Guangxi/CN | JX241046.1 | 78.9% | 88.0% | 87.5% | 81.9% | 82.6% |
| 46 | GX1105 | 2012 | Guangxi/CN | JX241045.1 | 78.9% | 88.0% | 87.7% | 82.3% | 82.7% |
| Strain | Nucleotide and Deduced Amino Acid Lengths of Region in Isolated DuCVs | Identities of Nucleotide and Deduced Amino Acid (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cap nt (aa) | Rep nt (aa) | Full Genome (nt) | DB2022-1 | DB2022-2 | DB2022-3 | DB2022-4 | |||||
| Cap nt (aa) | Rep nt (aa) | Cap nt (aa) | Rep nt (aa) | Cap nt (aa) | Rep nt (aa) | Cap nt (aa) | Rep nt (aa) | ||||
| DB2022-1 | 774 (257) | 879 (292) | 1993 | - | - | ||||||
| DB2022-2 | 774 (257) | 879 (292) | 1993 | 99.7 (100) | 100 (100) | ||||||
| DB2022-3 | 774 (257) | 879 (292) | 1993 | 99.7 (100) | 100 (100) | 100 (100) | 100 (100) | ||||
| DB2022-4 | 774 (257) | 879 (292) | 1994 | 99.7 (100) | 100 (100) | 100 (100) | 100 (100) | 100 (100) | 100 (100) | - | - |
| No | Strain | Year | Region | Accession No. | Genotype | Identities of Full Genome Sequence (%) | |
|---|---|---|---|---|---|---|---|
| nt | aa | ||||||
| 1 | VF-10 | 2012 | Shandong/CN | KC191683.1 | DHAV-3 | 98.2% | 99.6% |
| 2 | YT-57 | 2012 | Shandong/CN | KC191691.1 | DHAV3 | 98.6% | 100.0% |
| 3 | YA140902 | 2016 | Sichuan/CN | KX523288.1 | DHAV3 | 98.3% | 100.0% |
| 4 | ZP | 2014 | Shandong/CN | KM267028.1 | DHAV3 | 98.5% | 100.0% |
| 5 | GY140703 | 2016 | Sichuan/CN | KX523285.1 | DHAV3 | 98.2% | 99.6% |
| 6 | SD151227 | 2016 | Sichuan/CN | KX523287.1 | DHAV3 | 98.1% | 100.0% |
| 7 | A/dk/CHN/SD08/2019 | 2020 | Shandong/CN | MN912703.1 | DHAV3 | 97.9% | 99.6% |
| 8 | JS | 2020 | Heilongjiang/CN | MN164467.1 | DHAV3 | 99.7% | 100.0% |
| 9 | QL150713 | 2016 | Sichuan/CN | KX523286.1 | DHAV3 | 98.3% | 100.0% |
| 10 | YA151006 | 2016 | Sichuan/CN | KX523289.1 | DHAV3 | 98.5% | 100.0% |
| 11 | JN-12 | 2012 | Shandong/CN | KC191689.1 | DHAV3 | 97.2% | 99.6% |
| 12 | C-LX | 2009 | Beijing/CN | FJ626667.1 | DHAV3 | 97.4% | 99.2% |
| 13 | VF-40 | 2012 | Shandong/CN | KC191686.1 | DHAV3 | 96.7% | 98.3% |
| 14 | C-PSY | 2009 | Beijing/CN | FJ626670.1 | DHAV3 | 96.8% | 98.3% |
| 15 | C-YCZ | 2009 | Beijing/CN | FJ626672.1 | DHAV3 | 96.5% | 97.9% |
| 16 | YT-63 | 2012 | Shandong/CN | KC191693.1 | DHAV3 | 94.0% | 95.4% |
| 17 | BM | 2011 | Vietnam | JF925119.1 | DHAV3 | 95.8% | 97.5% |
| 18 | C-YZC | 2009 | Beijing/CN | FJ626673.1 | DHAV3 | 95.7% | 97.1% |
| 19 | C-PJK | 2012 | Sichuan/CN | KC282430.1 | DHAV3 | 94.0% | 96.2% |
| 20 | HY | 2014 | Vietnam | KM361878.1 | DHAV3 | 95.1% | 97.5% |
| 21 | GD1 | 2007 | Jiangsu/CN | EU289393.1 | DHAV3 | 94.7% | 96.7% |
| 22 | YT-BX | 2012 | Shandong/CN | KC191694.1 | DHAV3 | 94.2% | 95.4% |
| 23 | NT | 2014 | Vietnam | KM361883.1 | DHAV3 | 94.2% | 95.8% |
| 24 | NC | 2011 | Vietnam | JF925121.1 | DHAV3 | 91.0% | 92.1% |
| 25 | DN2 | 2014 | Vietnam | KM361877.1 | DHAV3 | 91.0% | 92.1% |
| 26 | KHO2 | 2014 | Vietnam | KM361880.1 | DHAV3 | 90.6% | 90.8% |
| 27 | LA1 | 2014 | Vietnam | KM361881.1 | DHAV3 | 90.6% | 90.4% |
| 28 | SY1 | 2007 | Zhejiang/CN | EF407857.1 | DHAV1 | 70.0% | 76.7% |
| 29 | ZI07-3 | 2007 | Zhejiang/CN | EF502170.1 | DHAV1 | 69.9% | 75.8% |
| 30 | SY2 | 2007 | Zhejiang/CN | EF407858.1 | DHAV1 | 70.2% | 76.7% |
| 31 | ZJ0812 | 2011 | Shanghai/CN | JF437541.1 | DHAV1 | 69.6% | 75.8% |
| 32 | SY6 | 2007 | Zhejiang/CN | EF407862.1 | DHAV1 | 70.4% | 76.7% |
| 33 | C-GA | 2008 | Beijing/CN | EU687759.1 | DHAV1 | 70.2% | 75.8% |
| 34 | C-TX | 2008 | Beijing/CN | EU687760.1 | DHAV1 | 70.0% | 75.8% |
| 35 | C-PHG | 2009 | Beijing/CN | FJ626668.1 | DHAV1 | 70.4% | 75.8% |
| 36 | C-QDY | 2009 | Beijing/CN | FJ626671.1 | DHAV1 | 70.7% | 75.8% |
| 37 | C-YXQ | 2008 | Beijing/CN | EU687762.1 | DHAV1 | 70.6% | 75.8% |
| 38 | JX140 | 2008 | Beijing/CN | KT072750.1 | DHAV1 | 71.1% | 76.7% |
| 39 | GD06 | 2018 | Fujian/CN | MH778934.1 | DHAV1 | 71.0%% | 76.7% |
| 40 | CZ141017 | 2016 | Sichuan/CN | KX523281.1 | DHAV1 | 70.4% | 76.7% |
| 41 | DY140903 | 2016 | Sichuan/CN | KX523282.1 | DHAV1 | 71.0% | 76.2% |
| 42 | C-CQ3 | 2008 | Beijing/CN | EU687761.1 | DHAV1 | 70.4% | 76.7% |
| 43 | LY0801-P30 | 2014 | Shandong/CN | KJ524552.1 | DHAV1 | 69.0% | 77.5% |
| 44 | MY | 2009 | Sichuan/CN | GU363950.1 | DHAV1 | 71.3% | 77.1% |
| 45 | LY1903 | 2019 | Shandong/CN | MN319537.1 | DHAV1 | 70.2% | 75.4% |
| 46 | SQ1903 | 2019 | Shandong/CN | MN319539.1 | DHAV1 | 70.6% | 75.8% |
| 47 | SY0812 | 2011 | Shanghai/CN | JF437538.1 | DHAV1 | 70.0% | 75.0% |
| 48 | LY0801-CP20 | 2014 | Shandong/CN | KJ606043.1 | DHAV1 | 70.3% | 75.4% |
| 49 | LY0801-CP30 | 2014 | Shandong/CN | KJ606045.1 | DHAV1 | 70.3% | 75.4% |
| No | Strain | Year | Region | Accession No. | Genotype | Whole-Genome Sequence Identity (%) | |
|---|---|---|---|---|---|---|---|
| nt | aa | ||||||
| 1 | HB06 | 2008 | Beijing/CN | EU526387.1 | I | 98.3% | 96.3% |
| 2 | XJ-2011-0315 | 2013 | Heilongjiang/CN | KC963045.1 | I | 98.5% | 97.2% |
| 3 | JR1 | 2006 | USA | EF122836.1 | I | 98.8% | 97.9% |
| 4 | LN09-1 | 2014 | Beijing/CN | KP288843.1 | I | 98.8% | 97.9% |
| 5 | HB10-1 | 2014 | Beijing/CN | KP288833.1 | I | 98.8% | 97.6% |
| 6 | BJ10-1 | 2014 | Beijing/CN | KP288867.1 | I | 98.3% | 97.9% |
| 7 | S113 | 1995 | Canada | L39002.1 | I | 98.2% | 96.3% |
| 8 | 601SI | 1999 | Taiwan/CN | AF204947.1 | I | 98.0% | 96.3% |
| 9 | 1733 | 1997 | USA | AF004857.1 | I | 99.4% | 98.8% |
| 10 | GuangxiR1 | 2012 | Guangxi/CN | KC183744.1 | I | 99.3% | 98.5% |
| 11 | GuangxiR2 | 2013 | Guangxi/CN | KF741732.1 | I | 99.3% | 98.5% |
| 12 | Fahey-Crawley | 2006 | Brazil | DQ868789.1 | I | 99.5% | 99.1% |
| 13 | 176 | 1999 | Canada | AF218358.1 | I | 99.5% | 99.1% |
| 14 | G-98 | 2006 | Huhhot/CN | DQ643975.1 | I | 99.7% | 99.7% |
| 15 | 2012-0129 | 2013 | Heilongjiang/CN | KC963052.1 | I | 99.7% | 99.7% |
| 16 | ARV-HeB01 | 2016 | Heilongjiang/CN | KX451231.1 | I | 99.7% | 99.7% |
| 17 | ARV-XY01 | 2016 | Heilongjiang/CN | KX451229.1 | I | 99.5% | 99.1% |
| 18 | SD-2010-0085 | 2013 | Heilongjiang/CN | KC963039.1 | I | 99.4% | 98.8% |
| 19 | LN05 | 2016 | Heilongjiang/CN | KX451225.1 | I | 99.7% | 99.7% |
| 20 | C-98 | 2006 | Huhhot/CN | EF057397.1 | I | 99.6% | 99.4% |
| 21 | T-98 | 2006 | Huhhot/CN | EF057398.1 | I | 99.5% | 99.4% |
| 22 | ARV-LN01 | 2016 | Heilongjiang/CN | KX451221.1 | I | 99.6% | 99.4% |
| 23 | ARV-LN06 | 2016 | Heilongjiang/CN | KX451226.1 | I | 99.6% | 99.4% |
| 24 | B-98 | 2006 | Huhhot/CN | DQ643974.1 | I | 99.5% | 99.1% |
| 25 | VA-1 | 2008 | India | EU681254.1 | I | 98.9% | 97.2% |
| 26 | GEL12 98M | 2001 | Netherlands | AF354225.1 | I | 77.3% | 77.6% |
| 27 | Reo/Broiler/YTLY/161024a | 2018 | Shandong/CN | MK189468.1 | I | 73.7% | 73.9% |
| 28 | 916 | 2000 | Taiwan/CN | AF297214.1 | II | 61.2% | 58.1% |
| 29 | ARV/Crow/Kagawa/P3/2012 | 2016 | Japan | LC164026.1 | II | 61.0% | 56.6% |
| 30 | GEL13a98M | 2001 | Netherlands | AF354226.1 | II | 51.3% | 54.4% |
| 31 | NC/98 | 2006 | USA | DQ995806.1 | II | 61.0% | 55.7% |
| 32 | TARV-MN3 | 2013 | USA | KF872234.1 | II | 53.3% | 56.7% |
| 33 | TARV-ONEIL | 2013 | USA | KF872231.1 | II | 52.0% | 56.2% |
| 34 | TARV-Crestview | 2013 | USA | KF872238.1 | II | 52.5% | 55.5% |
| 35 | T1781 | 2013 | Hungary | KC865792.1 | III | 59.7% | 50.8% |
| 36 | SD18 | 2020 | Shandong/CN | MT747423.1 | III | 59.3% | 50.5% |
| 37 | 42563-4 | 2006 | USA | DQ872801.1 | III | 58.9% | 54.7% |
| 38 | GEL13b98M | 2001 | Netherlands | AF354227.1 | III | 58.1% | 53.5% |
| 39 | D6 | 2019 | Canada | MN879650.1 | IV | 45.6% | 48.9% |
| 40 | D12 | 2019 | Canada | MN879710.1 | IV | 55.5% | 49.8% |
| 41 | Reo/USA/Broiler/1057NY/18 | 2021 | USA | MW854823.1 | IV | 56.2% | 49.5% |
| 42 | AVS-B | 2010 | Hungary | FR694197.1 | IV | 54.8% | 48.6% |
| 43 | K1600657 | 2019 | USA | MK583337.1 | IV | 54.7% | 49.5% |
| 44 | NLI12 96M | 2001 | Netherlands | AF354230.1 | IV | 54.1% | 50.2% |
| 45 | 918 | 2000 | Taiwan/CN | AF297215.1 | V | 57.0% | 48.0% |
| 46 | 1017-1 | 2000 | Taiwan/CN | AF297216.1 | V | 56.5% | 48.3% |
| 47 | NGN20 7-1 2b | 2021 | Japan | LC604650.1 | V | 55.9% | 47.0% |
| 48 | Reo/PA/Broiler/19981/13 | 2015 | USA | KR856993.1 | VI | 55.5% | 49.3% |
| 49 | Reo/PA/Broiler/03476/12 | 2015 | USA | KP727784.1 | VI | 55.8% | 48.0% |
| 50 | Reo/PA/Broiler/03200/12 | 2015 | USA | KP727785.1 | VI | 55.8% | 48.0% |
| Sequence | pI | Instability Index | Aliphatic Index | GRAVY |
|---|---|---|---|---|
| DuCV-KC851817 (2013) | 10.74 | 58.9 | 50.47 | −0.748 |
| DuCV-OQ658501.1 (2022) | 10.61 | 60.94 | 50.08 | −0.765 |
| DHAV-3-FJ626673 (2009) | 6.62 | 51.32 | 83.67 | −0.149 |
| DHAV-3-OQ658504.1 (2022) | 7.29 | 52.39 | 82.04 | −0.163 |
| ARV-EF122836 (2006) | 5.0 | 41.68 | 89.72 | −0.057 |
| ARV-OR046324.1 (2022) | 4.81 | 36.12 | 89.11 | −0.036 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Yu, H.; Zhang, W.; Fu, L.; Wang, Y. Molecular Detection and Genetic Characterization of Vertically Transmitted Viruses in Ducks. Animals 2024, 14, 6. https://doi.org/10.3390/ani14010006
Wang X, Yu H, Zhang W, Fu L, Wang Y. Molecular Detection and Genetic Characterization of Vertically Transmitted Viruses in Ducks. Animals. 2024; 14(1):6. https://doi.org/10.3390/ani14010006
Chicago/Turabian StyleWang, Xinrong, Haidong Yu, Wenli Zhang, Lizhi Fu, and Yue Wang. 2024. "Molecular Detection and Genetic Characterization of Vertically Transmitted Viruses in Ducks" Animals 14, no. 1: 6. https://doi.org/10.3390/ani14010006
APA StyleWang, X., Yu, H., Zhang, W., Fu, L., & Wang, Y. (2024). Molecular Detection and Genetic Characterization of Vertically Transmitted Viruses in Ducks. Animals, 14(1), 6. https://doi.org/10.3390/ani14010006







