Integrated miRNA and mRNA Sequencing Reveals the Sterility Mechanism in Hybrid Yellow Catfish Resulting from Pelteobagrus fulvidraco (♀) × Pelteobagrus vachelli (♂)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Ethics Statements
2.2. Extraction of Total RNA and Transcriptome Sequencing
2.3. Small-RNA Library Sequencing and Analysis
2.4. GO, KEGG, and HP Enrichment Analyses
2.5. Prediction of microRNA Target Genes and Differential Expression of Known miRNAs
2.6. Real-Time Quantitative PCR (qPCR)
2.7. Statistical Analysis
3. Results
3.1. Data Collection and Transcriptome Assembly
3.2. Identification of DEGs between Yellow Catfish Species
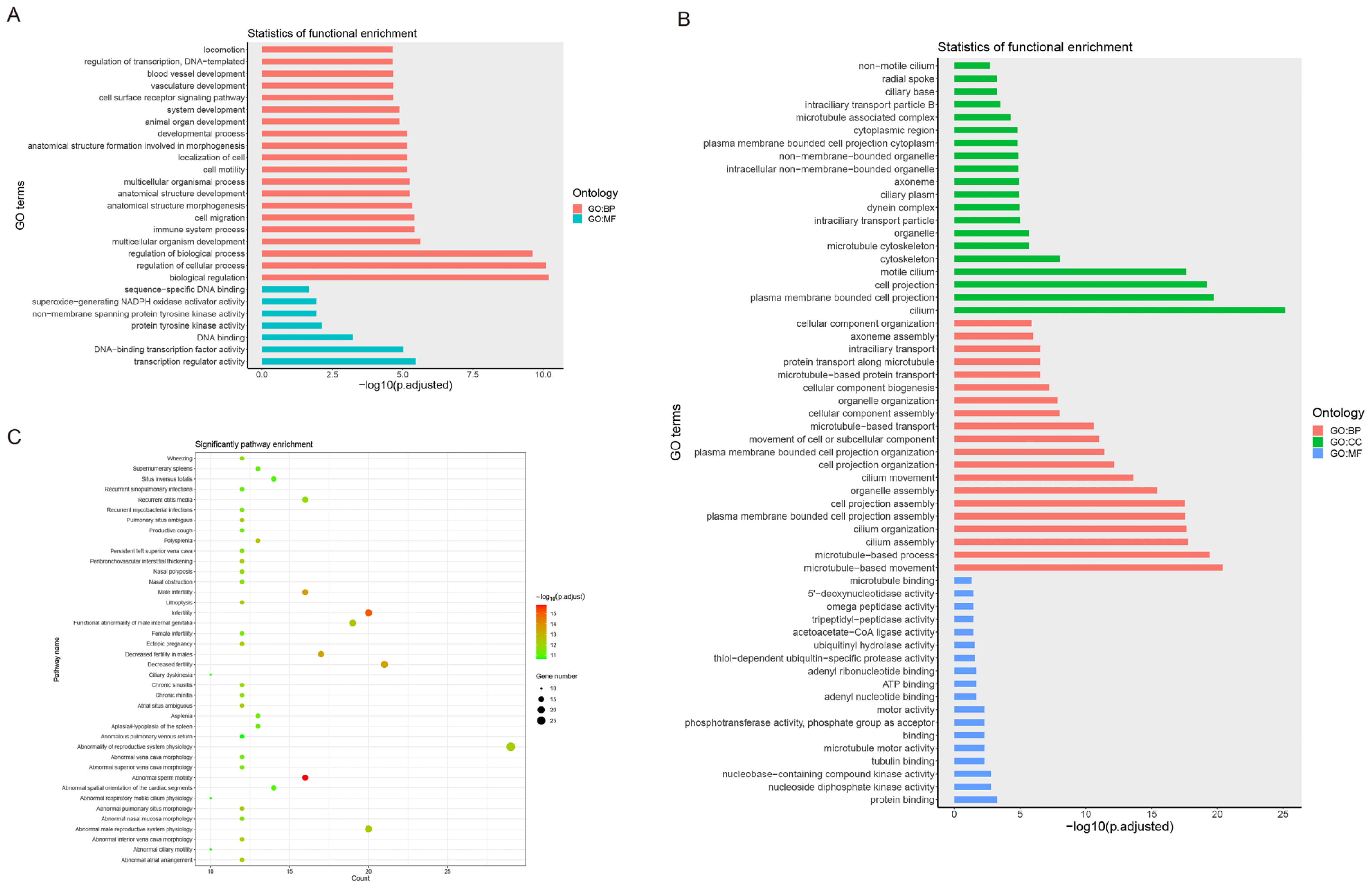
3.3. Enrichment Analysis of DEGs
3.4. Identification of miRNAs
3.5. Annotation of the Known miRNAs
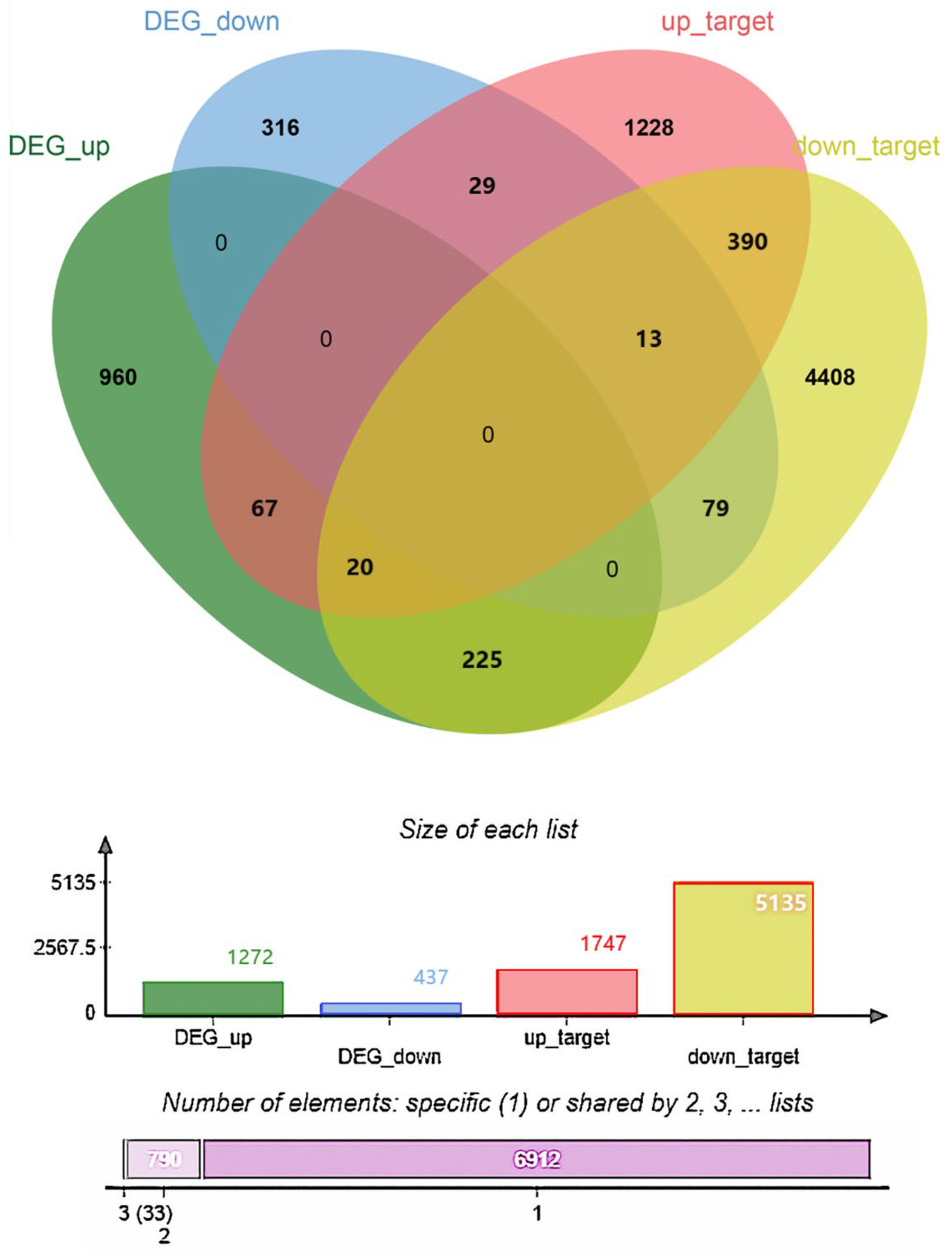
3.6. Identification of DEmiRNA and Enrichment Analyses of Target Genes of DEmiRNAs
3.7. Identification of DEmiRNAs and Enrichment Analyses of Target Genes of DEmiRNAs
3.8. RT-qPCR Verification
4. Discussion
4.1. DEGs between Hybrid and Pure Yellow Catfishes
4.2. Sexual Dimorphism in Hybrid Sterility of Yellow Catfish
4.3. Functional Enrichment of DEGs in Hybrid Yellow Catfishes
4.4. Expression Validation Using qRT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Z.H.; Chen, Q.L.; Chen, Q.; Li, F.; Li, Y.W. Diethylstilbestrol Arrested Spermatogenesis and Somatic Growth in the Juveniles of Yellow Catfish (Pelteobagrus fulvidraco), a Fish with Sexual Dimorphic Growth. Fish Physiol. Biochem. 2018, 44, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yang, X.; Shao, C.; Wang, Y.; Wang, X.; Mu, L.; Lv, J.; Wang, J. Complete Mitochondrial Genome of the Hybrid Loach of Paramisgurnus dabryanus ssp. (Female) and Misgurnus bipartitus (Male). Mitochondrial DNA Part B 2019, 4, 1727–1728. [Google Scholar] [CrossRef]
- Huang, J.; Wen, H.C.; Shi, J.G.; Huang, Y.H.; Liang, Z.S.; Long, Y.N.; Qin, J.X. Isolation and Identification of Pathogenic Bacteria from Pelteobagrus fulvidraco Ulcerative Syndrome and Its Drug Sensitive Test. J. South. Agric. 2012, 43, 107–112. [Google Scholar]
- Gong, G.; Dan, C.; Xiao, S.; Guo, W.; Huang, P.; Xiong, Y.; Wu, J.; He, Y.; Zhang, J.; Li, X.; et al. Chromosomal-Level Assembly of Yellow Catfish Genome Using Third-Generation DNA Sequencing and Hi-C Analysis. GigaScience 2018, 7, giy120. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yin, S.; Wang, Y.; Li, L.; Wang, X.; Ding, Y.; Zang, X.; Zhang, H.; Jia, Y.; Hu, Y. The Effects of Water Temperature and Stocking Density on Survival, Feeding and Growth of the Juveniles of the Hybrid Yellow Catfish from Pelteobagrus fulvidraco (♀) × Pelteobagrus vachelli (♂). Aquac. Res. 2016, 47, 2844–2850. [Google Scholar] [CrossRef]
- Qin, C.; Gong, Q.; Wen, Z.; Zou, Y.; Yuan, D.; Shao, T.; Li, H. Comparative Analysis of the Liver Transcriptome of Pelteobagrus vachellii with an Alternative Feeding Time. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 22, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Lou, Y. Fish Breeding Science; China Agriculture Press: Beijing, China, 1999. (In Chinese) [Google Scholar]
- Bartley, D.M.; Rana, K.J.; Immink, A. The Use of Inter-Specific Hybrids in Aquaculture and Fisheries. Rev. Fish Biol. Fish. 2001, 10, 325–337. [Google Scholar] [CrossRef]
- Zhang, G.; Li, J.; Zhang, J.; Liang, X.; Zhang, X.; Wang, T.; Yin, S. Integrated Analysis of Transcriptomic, miRNA and Proteomic Changes of a Novel Hybrid Yellow Catfish Uncovers Key Roles for miRNAs in Heterosis. Mol. Cell. Proteom. 2019, 18, 1437–1453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tao, P.; Chen, J.; Wang, R.; Zang, X.; Yin, S. The Complete Mitochondrial Genome of the Hybrid of Pelteobagrus fulvidraco (♀) × Pelteobagrus vachelli (♂). Mitochondrial DNA Part DNA Mapp. Seq. Anal. 2016, 27, 4191–4192. [Google Scholar] [CrossRef]
- Hu, W. Studies on the Genetic Characteristics of Yellow Catfish “Huang You No. 1” and Improvement of Female Reproductive Performance. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2020. [Google Scholar]
- Hu, W.; Chu, Z.; Xiong, Y.; Mei, J.; Lin, Q.; Ren, F.; Huang, P.; Guo, W. Defective germplasm assembly and germ cell development contribute to hybrid sterility in yellow catfish. Aquac. Res. 2021. [Google Scholar] [CrossRef]
- Xie, Y.; Shen, R.; Chen, L.; Liu, Y.G. Molecular Mechanisms of Hybrid Sterility in Rice. Sci. China Life Sci. 2019, 62, 737–743. [Google Scholar] [CrossRef]
- Li, J.; Zhou, J.; Zhang, Y.; Yang, Y.; Pu, Q.; Tao, D. New Insights into the Nature of Interspecific Hybrid Sterility in Rice. Front. Plant Sci. 2020, 11, 555572. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Wu, S.; Li, Z.; Dong, Z.; An, X.; Ma, B.; Tian, Y.; Li, J. Maize Genic Male-Sterility Genes and Their Applications in Hybrid Breeding: Progress and Perspectives. Mol. Plant 2019, 12, 321–342. [Google Scholar] [CrossRef] [PubMed]
- Milner, M.J.; Craze, M.; Bowden, S.; Bates, R.; Wallington, E.J.; Keeling, A. Identification of Genes Involved in Male Sterility in Wheat (Triticum aestivum L.) Which Could Be Used in a Genic Hybrid Breeding System. Plant Direct 2020, 4, e00201. [Google Scholar] [CrossRef]
- Niayale, R.; Cui, Y.; Adzitey, F. Male Hybrid Sterility in the Cattle-Yak and Other Bovines: A Review. Biol. Reprod. 2021, 104, 495–507. [Google Scholar] [CrossRef]
- Alhazmi, D.; Fudyk, S.K.; Civetta, A. Testes Proteases Expression and Hybrid Male Sterility Between Subspecies of Drosophila pseudoobscura. G3 Genes|Genomes|Genet. 2019, 9, 1065–1074. [Google Scholar] [CrossRef]
- Xu, D.; Yoshino, T.; Konishi, J.; Yoshikawa, H.; Ino, Y.; Yazawa, R.; Dos Santos Nassif Lacerda, S.M.; de França, L.R.; Takeuchi, Y. Germ Cell-Less Hybrid Fish: Ideal Recipient for Spermatogonial Transplantation for the Rapid Production of Donor-Derived Sperm. Biol. Reprod. 2019, 101, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, H.; Xu, D.; Ino, Y.; Yoshino, T.; Hayashida, T.; Wang, J.; Yazawa, R.; Yoshizaki, G.; Takeuchi, Y. Hybrid Sterility in Fish Caused by Mitotic Arrest of Primordial Germ Cells. Genetics 2018, 209, 507–521. [Google Scholar] [CrossRef] [PubMed]
- Hale, D.W.; Washburn, L.L.; Eicher, E.M. Meiotic Abnormalities in Hybrid Mice of the C57BL/6J x Mus Spretus Cross Suggest a Cytogenetic Basis for Haldane’s Rule of Hybrid Sterility. Cytogenet. Cell Genet. 1993, 63, 221–234. [Google Scholar] [CrossRef]
- Kidwell, M.G.; Kidwell, J.F.; Sved, J.A. Hybrid Dysgenesis in Drosophila melanogaster: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics 1977, 86, 813–833. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Bikard, D.; Patel, D.; Le Metté, C.; Giorgi, V.; Camilleri, C.; Bennett, M.J.; Loudet, O. Divergent Evolution of Duplicate Genes Leads to Genetic Incompatibilities within A. Thaliana. Science 2009, 323, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Chou, J.Y.; Cheong, L.; Chang, N.H.; Yang, S.Y.; Leu, J.Y. Incompatibility of Nuclear and Mitochondrial Genomes Causes Hybrid Sterility between Two Yeast Species. Cell 2008, 135, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Isolating Mechanisms, Evolution and Temperature. Biol. Symp. 1942, 6, 71. [Google Scholar]
- Rand, D.M.; Haney, R.A.; Fry, A.J. Cytonuclear Coevolution: The Genomics of Cooperation. Trends Ecol. Evol. 2004, 19, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.H.; Dan, C.; Guo, W.J.; Fan, Q.X.; Mei, J. The Morphology and Gonad Development of Pelteobagrus fulvidraco and Its Interspecific Hybrid “Huangyou No. 1” with Pelteobaggrus vachelli. Acta Hydrobiol. Sin. 2019, 43, 1231–1238. [Google Scholar]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Bao, Z.; Yang, M.; Shen, Y.; Zhang, X.; Yue, B.; Meng, Y.; Fan, Z. Identification and Characterization of microRNAs in American Cockroach (Periplaneta americana). Gene 2020, 743, 144610. [Google Scholar] [CrossRef]
- Yang, Q.; Yu, J.; Jiang, L.; Liu, X.; Liu, F.; Cai, Y.; Niu, L.; Price, M.; Li, J. Identification and Expression Profile of microRNA in Seven Tissues of the Golden Snub-Nosed Monkey (Rhinopithecus roxellanae). Mol. Genet. Genom. 2020, 295, 1547–1558. [Google Scholar] [CrossRef]
- Asgari, S. MicroRNA Functions in Insects. Insect Biochem. Mol. Biol. 2013, 43, 388–397. [Google Scholar] [CrossRef]
- Cui, J.; Zhou, B.; Ross, S.A.; Zempleni, J. Nutrition, microRNAs, and Human Health. Adv. Nutr. 2017, 8, 105–112. [Google Scholar] [CrossRef]
- Ha, M.; Lu, J.; Tian, L.; Ramachandran, V.; Kasschau, K.D.; Chapman, E.J.; Carrington, J.C.; Chen, X.; Wang, X.J.; Chen, Z.J. Small RNAs Serve as a Genetic Buffer against Genomic Shock in Arabidopsis Interspecific Hybrids and Allopolyploids. Proc. Natl. Acad. Sci. USA 2009, 106, 17835–17840. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-Genome Sequence of a Flatfish Provides Insights into ZW Sex Chromosome Evolution and Adaptation to a Benthic Lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, F.; Bao, W.; He, J.; Li, X.; Chen, J.; Xu, P. Responses of Functional miRNA-mRNA Regulatory Modules to a High-Fat Diet in the Liver of Hybrid Yellow Catfish (Pelteobagrus fulvidraco × P. vachelli). Genomics 2021, 113, 1207–1220. [Google Scholar] [CrossRef]
- Qiang, J.; Tao, F.; Bao, W.; He, J.; Liang, M.; Liang, C.; Zhu, H.; Li, X.; Chen, D.; Xu, P. miR-489-3p Regulates the Oxidative Stress Response in the Liver and Gill Tissues of Hybrid Yellow Catfish (Pelteobagrus fulvidraco♀ × P. vachelli♂) Under Cu2+ Exposure by Targeting Cu/Zn-SOD. Front. Physiol. 2019, 10, 868. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.G.; Zhang, Y.G.; He, S.P.; Chen, Y.Y. Phylogeny of chinese catfishes inferred from mitochondrial cytochrome b sequences. Yi Chuan Xue Bao 2005, 32, 145–154. (In Chinese) [Google Scholar] [PubMed]
- Zhang, X.; Hu, R.; Yang, Y. Comparative Histological Studies on the Spleen of Fish, Bullfrog, Chicken and Mouse. J. Guiyang Med. Coll. 2007, 32, 644–645. [Google Scholar]
- Patel, R.K.; Jain, M. NGSQC Toolkit: A Toolkit for Quality Control of next Generation Sequencing Data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 Accurately Identifies Known and Hundreds of Novel microRNA Genes in Seven Animal Clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. MiRanda application: Human microRNA targets. PLoS Biol. 2005, 2, e363. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Sai Lakshmi, S.; Agrawal, S. piRNABank: A Web Resource on Classified and Clustered Piwi-Interacting RNAs. Nucleic Acids Res. 2008, 36, D173–D177. [Google Scholar] [CrossRef] [PubMed]
- Aprea, I.; Wilken, A.; Krallmann, C.; Nöthe-Menchen, T.; Olbrich, H.; Loges, N.T.; Dougherty, G.W.; Bracht, D.; Brenker, C.; Kliesch, S.; et al. Pathogenic Gene Variants in CCDC39, CCDC40, RSPH1, RSPH9, HYDIN, and SPEF2 Cause Defects of Sperm Flagella Composition and Male Infertility. Front. Genet. 2023, 14, 1117821. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Liang, Y.; Liu, M.; Yang, Y.; Liu, H.; Shen, Y. Novel Biallelic Mutations in TTC29 Cause Asthenoteratospermia and Male Infertility. Mol. Genet. Genom. Med. 2022, 10, e2078. [Google Scholar] [CrossRef] [PubMed]
- Shamoto, N.; Narita, K.; Kubo, T.; Oda, T.; Takeda, S. CFAP70 Is a Novel Axoneme-Binding Protein That Localizes at the Base of the Outer Dynein Arm and Regulates Ciliary Motility. Cells 2018, 7, 124. [Google Scholar] [CrossRef] [PubMed]
- Beurois, J.; Martinez, G.; Cazin, C.; Kherraf, Z.E.; Amiri-Yekta, A.; Thierry-Mieg, N.; Bidart, M.; Petre, G.; Satre, V.; Brouillet, S.; et al. CFAP70 Mutations Lead to Male Infertility Due to Severe Astheno-Teratozoospermia. A Case Report. Hum. Reprod. 2019, 34, 2071–2079. [Google Scholar] [CrossRef]
- Abbasi, F.; Miyata, H.; Shimada, K.; Morohoshi, A.; Nozawa, K.; Matsumura, T.; Xu, Z.; Pratiwi, P.; Ikawa, M. RSPH6A Is Required for Sperm Flagellum Formation and Male Fertility in Mice. J. Cell Sci. 2018, 131, jcs221648. [Google Scholar] [CrossRef]
- Luo, G.; Hou, M.; Wang, B.; Liu, Z.; Liu, W.; Han, T.; Zhang, D.; Zhou, X.; Jia, W.; Tan, Y.; et al. Tsga10 Is Essential for Arrangement of Mitochondrial Sheath and Male Fertility in Mice. Andrology 2021, 9, 368–375. [Google Scholar] [CrossRef]
- Kapanidou, M.; Curtis, N.L.; Bolanos-Garcia, V.M. Cdc20: At the Crossroads between Chromosome Segregation and Mitotic Exit. Trends Biochem. Sci. 2017, 42, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Kalous, J.; Aleshkina, D. Multiple Roles of PLK1 in Mitosis and Meiosis. Cells 2023, 12, 187. [Google Scholar] [CrossRef]
- Barreto, R.D.S.N.; Matias, G.D.S.S.; Nishiyama, M.Y., Jr.; Carreira, A.C.O.; Miglino, M.A. ECM Proteins Involved in Cell Migration and Vessel Formation Compromise Bovine Cloned Placentation. Theriogenology 2022, 188, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Fagegaltier, D.; König, A.; Gordon, A.; Lai, E.C.; Gingeras, T.R.; Hannon, G.J.; Shcherbata, H.R. A Genome-Wide Survey of Sexually Dimorphic Expression of Drosophila miRNAs Identifies the Steroid Hormone-Induced miRNA Let-7 as a Regulator of Sexual Identity. Genetics 2014, 198, 647–668. [Google Scholar] [CrossRef] [PubMed]
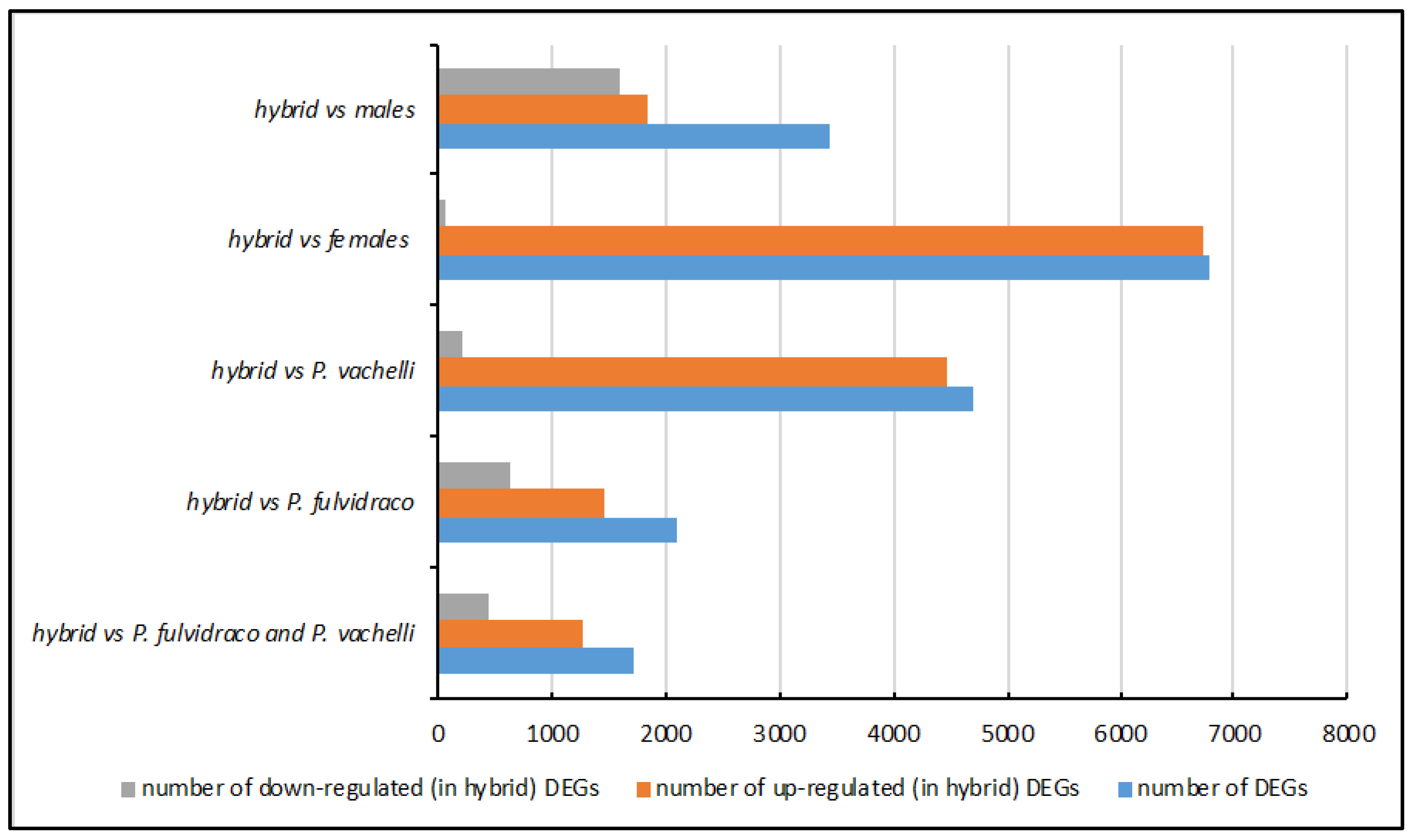

| Group | Number of DEGs | Number of Upregulated (in Hybrid) DEGs | Number of Downregulated (in Hybrid) DEGs |
|---|---|---|---|
| hybrid vs. P. fulvidraco and P. vachelli | 1709 | 1272 | 437 |
| hybrid vs. P. fulvidraco | 2093 | 1467 | 626 |
| hybrid vs. P. vachelli | 4694 | 4476 | 218 |
| hybrid vs. females | 6788 | 6715 | 63 |
| hybrid vs. males | 3444 | 1844 | 1600 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Yang, Q.; Li, M.; Lan, Y.; Song, Z. Integrated miRNA and mRNA Sequencing Reveals the Sterility Mechanism in Hybrid Yellow Catfish Resulting from Pelteobagrus fulvidraco (♀) × Pelteobagrus vachelli (♂). Animals 2024, 14, 1586. https://doi.org/10.3390/ani14111586
Li S, Yang Q, Li M, Lan Y, Song Z. Integrated miRNA and mRNA Sequencing Reveals the Sterility Mechanism in Hybrid Yellow Catfish Resulting from Pelteobagrus fulvidraco (♀) × Pelteobagrus vachelli (♂). Animals. 2024; 14(11):1586. https://doi.org/10.3390/ani14111586
Chicago/Turabian StyleLi, Shu, Qiao Yang, Maohua Li, Yue Lan, and Zhaobin Song. 2024. "Integrated miRNA and mRNA Sequencing Reveals the Sterility Mechanism in Hybrid Yellow Catfish Resulting from Pelteobagrus fulvidraco (♀) × Pelteobagrus vachelli (♂)" Animals 14, no. 11: 1586. https://doi.org/10.3390/ani14111586





