Mechanisms of Embryonic Stem Cell Pluripotency Maintenance and Their Application in Livestock and Poultry Breeding
Abstract
:Simple Summary
Abstract
1. Introduction
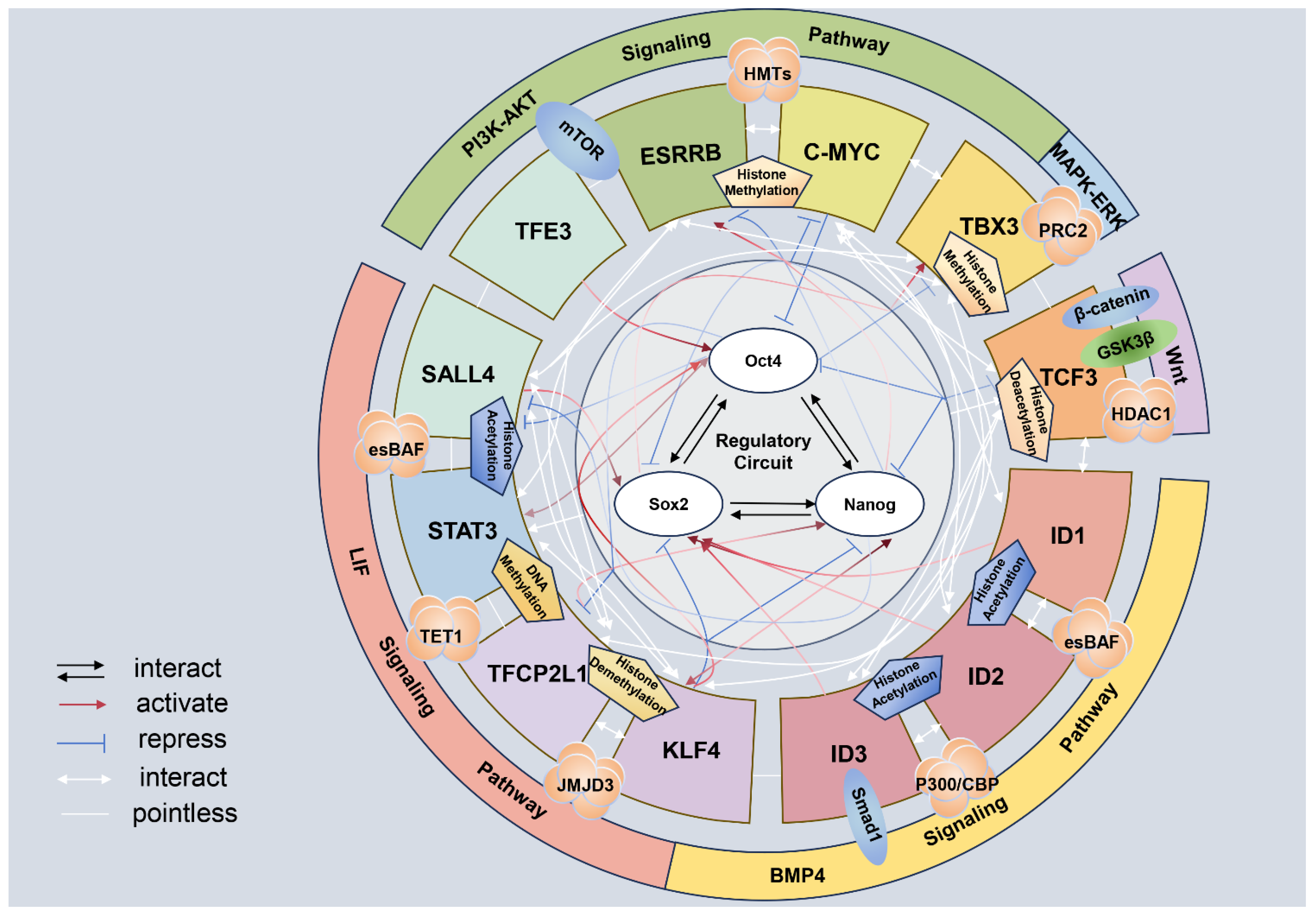
2. Mechanism of Pluripotency Maintenance of ESCs
2.1. Transcription Factor Related to Pluripotency
2.2. Signal Pathway
2.3. Epigenetic Modification
3. Application of Pluripotent Maintenance Mechanism (Induced Pluripotent Stem Cells)
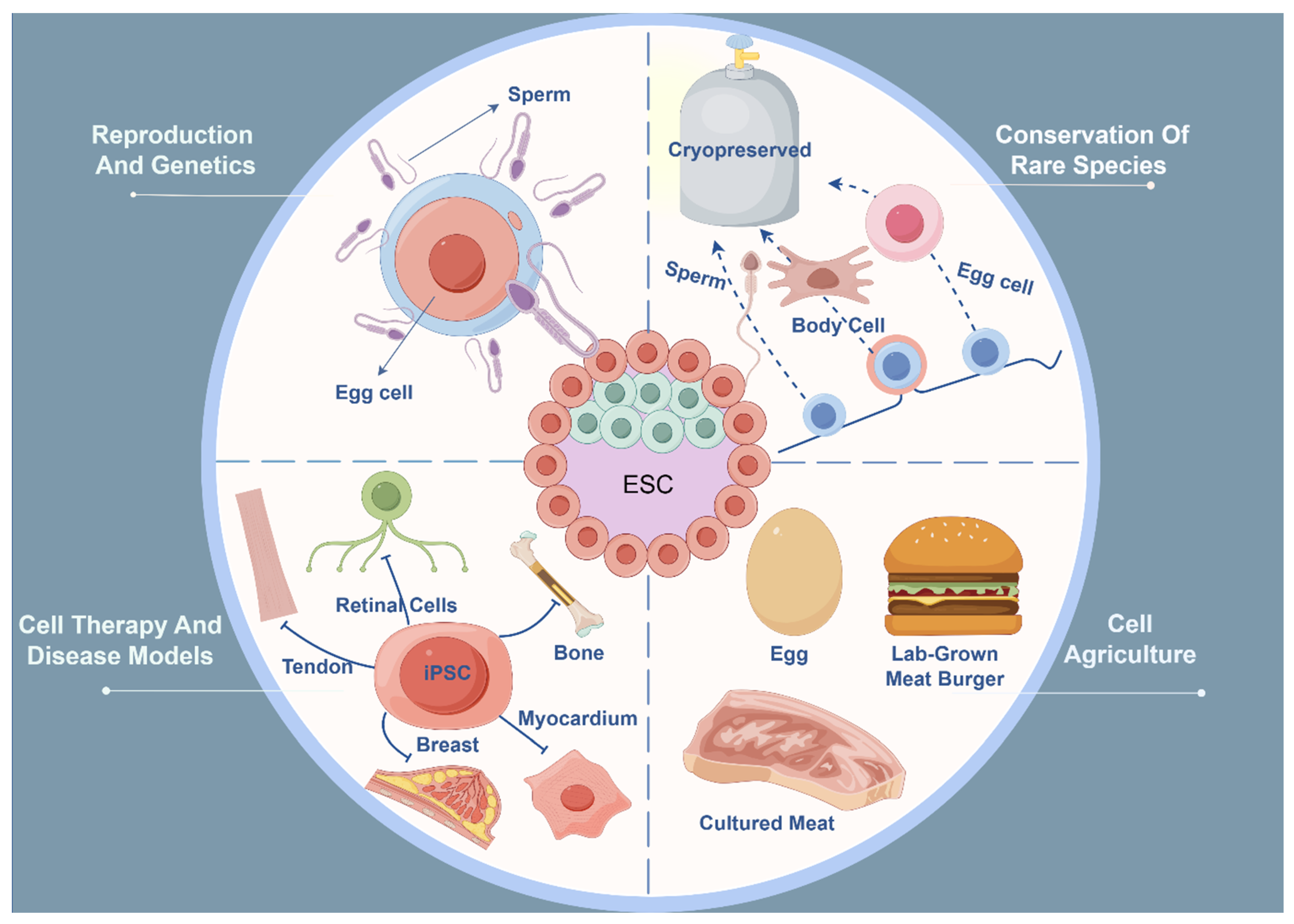
4. Application of Pluripotent Stem Cells
4.1. Reproduction: Theoretical Basis for the Application of Pluripotent Stem Cells
4.2. Conservation and Restoration of Rare Species (or Individuals)
4.3. Cell Agriculture (Cultured Meat, etc.)
4.4. Cell Therapy and Disease Models
5. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed]
- Thomson, A.J.; Pierart, H.; Meek, S.; Bogerman, A.; Sutherland, L.; Murray, H.; Mountjoy, E.; Downing, A.; Talbot, R.; Sartori, C.; et al. Reprogramming Pig Fetal Fibroblasts Reveals a Functional LIF Signaling Pathway. Cell. Reprogramming 2012, 14, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Boiani, M.; Scholer, H.R. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005, 6, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Varzideh, F.; Gambardella, J.; Kansakar, U.; Jankauskas, S.S.; Santulli, G. Molecular Mechanisms Underlying Pluripotency and Self-Renewal of Embryonic Stem Cells. Int. J. Mol. Sci. 2023, 24, 8386. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Sewell, W.; Lin, R.Y. Generation of thyroid follicular cells from pluripotent stem cells: Potential for regenerative medicine. Front. Endocrinol. 2014, 5, 96. [Google Scholar] [CrossRef] [PubMed]
- Posabella, A.; Alber, A.B.; Undeutsch, H.J.; Droeser, R.A.; Hollenberg, A.N.; Ikonomou, L.; Kotton, D.N. Derivation of Thyroid Follicular Cells From Pluripotent Stem Cells: Insights From Development and Implications for Regenerative Medicine. Front. Endocrinol. 2021, 12, 666565. [Google Scholar] [CrossRef]
- Su, Y.; Zhu, J.; Salman, S.; Tang, Y. Induced pluripotent stem cells from farm animals. J. Anim. Sci. 2020, 98, skaa343. [Google Scholar] [CrossRef]
- Loh, K.M.; Lim, B. A Precarious Balance: Pluripotency Factors as Lineage Specifiers. Cell Stem Cell 2011, 8, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Cheng, Y.Y.; Yen, C.Y.T.; Hsieh, P.C.H. Mechanisms of pluripotency maintenance in mouse embryonic stem cells. Cell. Mol. Life Sci. 2017, 74, 1805–1817. [Google Scholar] [CrossRef] [PubMed]
- Simandi, Z.; Horvath, A.; Wright, L.C.; Cuaranta-Monroy, I.; De Luca, I.; Karolyi, K.; Sauer, S.; Deleuze, J.F.; Gudas, L.J.; Cowley, S.M.; et al. OCT4 Acts as an Integrator of Pluripotency and Signal-Induced Differentiation. Mol. Cell 2016, 63, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Ambrosetti, D.C.; Schöler, H.R.; Dailey, L.; Basilico, C. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 2000, 275, 23387–23397. [Google Scholar] [CrossRef] [PubMed]
- Tapia, N.; MacCarthy, C.; Esch, D.; Marthaler, A.G.; Tiemann, U.; Araúzo-Bravo, M.J.; Jauch, R.; Cojocaru, V.; Schöler, H.R. Dissecting the role of distinct OCT4-SOX2 heterodimer configurations in pluripotency. Sci. Rep. 2015, 5, 13533. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.R.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.W.; Jiang, J.M.; et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.X.; Chao, C.; Saito, S.; Mazur, S.J.; Murphy, M.E.; Appella, E.; Xu, Y. P53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005, 7, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Loh, Y.H.; Wu, Q.; Chew, J.L.; Vega, V.B.; Zhang, W.W.; Chen, X.; Bourque, G.; George, J.; Leong, B.; Liu, J.; et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006, 38, 431–440. [Google Scholar] [CrossRef]
- Hanna, L.A.; Foreman, R.K.; Tarasenko, I.A.; Kessler, D.S.; Labosky, P.A. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Gene Dev. 2002, 16, 2650–2661. [Google Scholar] [CrossRef]
- Huang, G.Y.; Ye, S.D.; Zhou, X.L.; Liu, D.H.; Ying, Q.L. Molecular basis of embryonic stem cell self-renewal: From signaling pathways to pluripotency network. Cell. Mol. Life Sci. 2015, 72, 1741–1757. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, T.; Nakamura, T.; Nakao, K.; Arai, T.; Katsuki, M.; Heike, T.; Yokota, T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. Embo J. 1999, 18, 4261–4269. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Fujita, H.; Okada, Y.; Shinkai, S.; Onami, S.; Abe, K.; Fujimoto, K.; Sasaki, K.; Shioi, G.; Watanabe, T.M. Single-molecule tracking of Nanog and Oct4 in living mouse embryonic stem cells uncovers a feedback mechanism of pluripotency maintenance. Embo J. 2023, 42, e112305. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Ogawa, K.; Shimosato, D.; Adachi, K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature 2009, 460, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Masui, S. Pluripotency maintenance mechanism of embryonic stem cells and reprogramming. Int. J. Hematol. 2010, 91, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Niwa, H.; Burdon, T.; Chambers, I.; Smith, A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes. Dev. 1998, 12, 2048–2060. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Lee, D.S.; Yan, Y.T.; Shen, C.N.; Hwang, S.M.; Lee, S.T.; Hsieh, P.C. Bcl3 Bridges LIF-STAT3 to Oct4 Signaling in the Maintenance of Naive Pluripotency. Stem Cells 2015, 33, 3468–3480. [Google Scholar] [CrossRef] [PubMed]
- Mossahebi-Mohammadi, M.; Quan, M.Y.; Zhang, J.S.; Li, X.K. FGF Signaling Pathway: A Key Regulator of Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Bourillot, P.Y.; Savatier, P. Kruppel-like transcription factors and control of pluripotency. Bmc Biol. 2010, 8, 125. [Google Scholar] [CrossRef]
- Martello, G.; Bertone, P.; Smith, A. Identification of the missing pluripotency mediator downstream of leukaemia inhibitory factor. Embo J. 2013, 32, 2561–2574. [Google Scholar] [CrossRef]
- Jirmanova, L.; Afanassieff, M.; Gobert-Gosse, S.; Markossian, S.; Savatier, P. Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene 2002, 21, 5515–5528. [Google Scholar] [CrossRef] [PubMed]
- Burdon, T.; Stracey, C.; Chambers, I.; Nichols, J.; Smith, A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 1999, 210, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.N.; Totonchi, M.; Gourabi, H.; Schöler, H.R.; Baharvand, H. Signaling Roadmap Modulating Naive and Primed Pluripotency. Stem Cells Dev. 2014, 23, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.J.; Li, J.; Zhou, Y.L.; Zheng, H.; Pei, D.Q. A negative feedback loop of transcription factors that controls stem cell pluripotency and self-renewal. Faseb J. 2006, 20, 1730–1732. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.; Rendl, M.; Fuchs, E. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell 2006, 127, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.F.; Ng, D.Y.; Jayakumaran, G.; Wood, G.A.; Koide, H.; Doble, B.W. β-Catenin Enhances Oct-4 Activity and Reinforces Pluripotency through a TCF-Independent Mechanism. Cell Stem Cell 2011, 8, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Totty, N.F.; Irwin, M.S.; Sudol, M.; Downward, J. Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol. Cell 2003, 11, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Guan, K.L.; Figueroa, C.; Brtva, T.R.; Zhu, T.Q.; Taylor, J.; Barber, T.D.; Vojtek, A.B. Negative regulation of the serine/threonine kinase B-Raf by Akt. J. Biol. Chem. 2000, 275, 27354–27359. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Bio. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Duncan, E.J.; Gluckman, P.D.; Dearden, P.K. Epigenetics, plasticity, and evolution: How do we link epigenetic change to phenotype? J. Exp. Zool. B Mol. Dev. Evol. 2014, 322, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Shanak, S.; Helms, V. DNA methylation and the core pluripotency network. Dev. Biol. 2020, 464, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Olariu, V.; Lövkvist, C.; Sneppen, K. Nanog, Oct4 and Tet1 interplay in establishing pluripotency. Sci. Rep. 2016, 6, 25438. [Google Scholar] [CrossRef] [PubMed]
- Goszczynski, D.E.; Navarro, M.; Mutto, A.A.; Ross, P.J. Review: Embryonic stem cells as tools for in vitro gamete production in livestock. Animal 2023, 17 (Suppl. S1), 100828. [Google Scholar] [CrossRef] [PubMed]
- Swygert, S.G.; Peterson, C.L. Chromatin dynamics: Interplay between remodeling enzymes and histone modifications. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2014, 1839, 728–736. [Google Scholar] [CrossRef] [PubMed]
- van den Hurk, M.; Kenis, G.; Bardy, C.; van den Hove, D.L.; Gage, F.H.; Steinbusch, H.W.; Rutten, B.P. Transcriptional and epigenetic mechanisms of cellular reprogramming to induced pluripotency. Epigenomics 2016, 8, 1131–1149. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.; Avner, P. When X-inactivation meets pluripotency: An intimate rendezvous. FEBS Lett. 2009, 583, 1721–1727. [Google Scholar] [CrossRef] [PubMed]
- Ho, L.; Miller, E.L.; Ronan, J.L.; Ho, W.Q.; Jothi, R.; Crabtree, G.R. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat. Cell Biol. 2011, 13, 903–913. [Google Scholar] [CrossRef]
- Bai, J.; Xi, Q. Crosstalk between TGF-beta signaling and epigenome. Acta Biochim. Biophys. Sin. 2018, 50, 322. [Google Scholar] [CrossRef]
- Fagnocchi, L.; Mazzoleni, S.; Zippo, A. Integration of Signaling Pathways with the Epigenetic Machinery in the Maintenance of Stem Cells. Stem Cells Int. 2016, 2016, 8652748. [Google Scholar] [CrossRef]
- Lyu, J.; Jho, E.H.; Lu, W. Smek promotes histone deacetylation to suppress transcription of Wnt target gene brachyury in pluripotent embryonic stem cells. Cell Res. 2011, 21, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.F.; Johnstone, S.E.; Newman, J.J.; Kagey, M.H.; Young, R.A. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes. Dev. 2008, 22, 746–755. [Google Scholar] [CrossRef] [PubMed]
- Ying, Q.L.; Nichols, J.; Chambers, I.; Smith, A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 2003, 115, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, M.; Ueda, J.; Hayashi, K.; Ohta, H.; Yabuta, Y.; Kurimoto, K.; Nakato, R.; Yamada, Y.; Shirahige, K.; Saitou, M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell 2013, 12, 368–382. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.B.; Xi, Q.R. Crosstalk between TGF-β signaling and epigenome. Acta Bioch Bioph Sin. 2018, 50, 60–67. [Google Scholar] [CrossRef]
- Papatsenko, D.; Waghray, A.; Lemischka, I.R. Feedback control of pluripotency in embryonic stem cells: Signaling, transcription and epigenetics. Stem Cell Res. 2018, 29, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.L.; Ang, Y.S.; Sevilla, A.; Lemischka, I.R.; Ma’ayan, A. Construction and Validation of a Regulatory Network for Pluripotency and Self-Renewal of Mouse Embryonic Stem Cells. PLoS Comput. Biol. 2014, 10, e1003777. [Google Scholar] [CrossRef] [PubMed]
- Herberg, M.; Roeder, I. Computational modelling of embryonic stem-cell fate control. Development 2015, 142, 2250–2260. [Google Scholar] [CrossRef]
- Dunn, S.J.; Martello, G.; Yordanov, B.; Emmott, S.; Smith, A.G. Defining an essential transcription factor program for naive pluripotency. Science 2014, 344, 1156–1160. [Google Scholar] [CrossRef]
- Ray, A.; Joshi, J.M.; Sundaravadivelu, P.K.; Raina, K.; Lenka, N.; Kaveeshwar, V.; Thummer, R.P. An Overview on Promising Somatic Cell Sources Utilized for the Efficient Generation of Induced Pluripotent Stem Cells. Stem Cell Rev. Rep. 2021, 17, 1954–1974. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.Y.; Zhang, Y. Embryonic stem cell and induced pluripotent stem cell: An epigenetic perspective. Cell Res. 2013, 23, 49–69. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, M.; Zhou, H.Q.; Kasri, N.N. Choices for Induction of Pluripotency: Recent Developments in Human Induced Pluripotent Stem Cell Reprogramming Strategies. Stem Cell Rev. Rep. 2016, 12, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Meir, Y.J.J.; Li, G.G. Somatic Reprogramming-Above and Beyond Pluripotency. Cells 2021, 10, 2888. [Google Scholar] [CrossRef] [PubMed]
- Zahumenska, R.; Nosal, V.; Smolar, M.; Okajcekova, T.; Skovierova, H.; Strnadel, J.; Halasova, E. Induced Pluripotency: A Powerful Tool for In Vitro Modeling. Int. J. Mol. Sci. 2020, 21, 8910. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Wu, Z.; Wang, Y.; Cheng, L.; Cui, C.; Gao, Y.; Chen, T.; Rao, L.J.; Chen, S.Y.; Jia, N.N.; et al. Enhanced efficiency of generating induced pluripotent stem (iPS) cells from human somatic cells by a combination of six transcription factors. Cell Res. 2008, 18, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Zaehres, H.; Wu, G.M.; Gentile, L.; Ko, K.; Sebastiano, V.; Araúzo-Bravo, M.J.; Ruau, D.; Han, D.W.; Zenke, M.; et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 2008, 454, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Jiang, J.M.; Kraus, P.; Ng, J.H.; Heng, J.C.D.; Chan, Y.S.; Yaw, L.P.; Zhang, W.W.; Loh, Y.H.; Han, J.Y.; et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009, 11, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Sebastiano, V.; Wu, G.M.; Araúzo-Bravo, M.J.; Sasse, P.; Gentile, L.; Ko, K.; Ruau, D.; Ehrich, M.; van den Boom, D.; et al. Oct4-Induced Pluripotency in Adult Neural Stem Cells. Cell 2009, 136, 411–419. [Google Scholar] [CrossRef]
- Li, Y.Q.; Zhang, Q.A.; Yin, X.L.; Yang, W.F.; Du, Y.Y.; Hou, P.P.; Ge, J.A.; Liu, C.; Zhang, W.Q.; Zhang, X.; et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011, 21, 196–204. [Google Scholar] [CrossRef]
- Buganim, Y.; Markoulaki, S.; van Wietmarschen, N.; Hoke, H.; Wu, T.; Ganz, K.; Akhtar-Zaidi, B.; He, Y.P.; Abraham, B.J.; Porubsky, D.; et al. The Developmental Potential of iPSCs Is Greatly Influenced by Reprogramming Factor Selection. Cell Stem Cell 2014, 15, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Buganim, Y.; Faddah, D.A.; Cheng, A.W.; Itskovich, E.; Markoulaki, S.; Ganz, K.; Klemm, S.L.; van Oudenaarden, A.; Jaenisch, R. Single-Cell Expression Analyses during Cellular Reprogramming Reveal an Early Stochastic and a Late Hierarchic Phase. Cell 2012, 150, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.P.; Li, Y.Q.; Zhang, X.; Liu, C.; Guan, J.Y.; Li, H.G.; Zhao, T.; Ye, J.Q.; Yang, W.F.; Liu, K.; et al. Pluripotent Stem Cells Induced from Mouse Somatic Cells by Small-Molecule Compounds. Science 2013, 341, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, T.; Guan, J.Y.; Zhang, X.; Fu, Y.; Ye, J.Q.; Zhu, J.L.; Meng, G.F.; Ge, J.; Yang, S.S.; et al. A XEN-like State Bridges Somatic Cells to Pluripotency during Chemical Reprogramming. Cell 2015, 163, 1678–1691. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Huo, X.; Pei, G.; Jia, Z.; Yan, Y.; Yu, J.; Qu, H.; Xie, Y.; Yuan, J.; Zheng, Y.; et al. Dual-role transcription factors stabilize intermediate expression levels. Cell 2024, 187, 2746–2766.e25. [Google Scholar] [CrossRef]
- Liu, S.P.; Fu, R.H.; Huang, Y.C.; Chen, S.Y.; Chien, Y.J.; Hsu, C.Y.; Tsai, C.H.; Shyu, W.C.; Lin, S.Z. Induced pluripotent stem (iPS) cell research overview. Cell Transpl. 2011, 20, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Munis, A.M. Gene Therapy Applications of Non-Human Lentiviral Vectors. Viruses 2020, 12, 1106. [Google Scholar] [CrossRef] [PubMed]
- Chabanovska, O.; Galow, A.M.; David, R.; Lemcke, H. mRNA—A game changer in regenerative medicine, cell-based therapy and reprogramming strategies. Adv. Drug Deliv. Rev. 2021, 179, 114002. [Google Scholar] [CrossRef]
- Wang, S.; Qu, Z.; Huang, Q.; Zhang, J.; Lin, S.; Yang, Y.; Meng, F.; Li, J.; Zhang, K. Application of Gene Editing Technology in Resistance Breeding of Livestock. Life 2022, 12, 1070. [Google Scholar] [CrossRef] [PubMed]
- Pinzón-Arteaga, C.A.; Wang, Y.J.; Wei, Y.L.; Orsi, A.E.R.; Li, L.J.; Scatolin, G.; Liu, L.Z.; Sakurai, M.; Ye, J.F.; Ming, H.; et al. Bovine blastocyst-like structures derived from stem cell cultures. Cell Stem Cell 2023, 30, 611–616.e7. [Google Scholar] [CrossRef]
- Wang, H.; Xiang, J.; Zhang, W.; Li, J.; Wei, Q.; Zhong, L.; Ouyang, H.; Han, J. Induction of Germ Cell-like Cells from Porcine Induced Pluripotent Stem Cells. Sci. Rep. 2016, 6, 27256. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Feng, X.; Liao, S.; Zhang, D.; Cui, X.; Gao, F.; Han, C. Generation of male germ cells from mouse induced pluripotent stem cells in vitro. Stem Cell Res. 2014, 12, 517–530. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Wang, M.; Yuan, Y.; Wang, X.; Fu, R.; Wan, H.; Xie, M.; Liu, M.; Guo, X.; Zheng, Y.; et al. Complete Meiosis from Embryonic Stem Cell-Derived Germ Cells In Vitro. Cell Stem Cell 2016, 18, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Nowak-Imialek, M.; Chen, X.; Chen, D.; Herrmann, D.; Ruan, D.; Chen, A.C.H.; Eckersley-Maslin, M.A.; Ahmad, S.; Lee, Y.L.; et al. Establishment of porcine and human expanded potential stem cells. Nat. Cell Biol. 2019, 21, 687–699. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Castillo-Venzor, A.; Penfold, C.A.; Morgan, M.; Mizuno, N.; Tang, W.W.C.; Osada, Y.; Hirao, M.; Yoshida, F.; Sato, H.; et al. Tracing the emergence of primordial germ cells from bilaminar disc rabbit embryos and pluripotent stem cells. Cell Rep. 2021, 37, 109812. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wei, Y.; Sun, H.X.; Mahdi, A.K.; Pinzon Arteaga, C.A.; Sakurai, M.; Schmitz, D.A.; Zheng, C.; Ballard, E.D.; Li, J.; et al. Derivation of Intermediate Pluripotent Stem Cells Amenable to Primordial Germ Cell Specification. Cell Stem Cell 2021, 28, 550–567.e12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhi, M.; Gao, D.; Zhu, Q.; Gao, J.; Zhu, G.; Cao, S.; Han, J. Research progress and application prospects of stable porcine pluripotent stem cellsdagger. Biol. Reprod. 2022, 107, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Weeratunga, P.; Harman, R.M.; Van de Walle, G.R. Induced pluripotent stem cells from domesticated ruminants and their potential for enhancing livestock production. Front. Vet. Sci. 2023, 10, 1129287. [Google Scholar] [CrossRef]
- Hou, Z.; An, L.; Han, J.; Yuan, Y.; Chen, D.; Tian, J. Revolutionize livestock breeding in the future: An animal embryo-stem cell breeding system in a dish. J. Anim. Sci. Biotechnol. 2018, 9, 90. [Google Scholar] [CrossRef] [PubMed]
- Saragusty, J.; Diecke, S.; Drukker, M.; Durrant, B.; Friedrich Ben-Nun, I.; Galli, C.; Goritz, F.; Hayashi, K.; Hermes, R.; Holtze, S.; et al. Rewinding the process of mammalian extinction. Zoo. Biol. 2016, 35, 280–292. [Google Scholar] [CrossRef]
- Ben-Nun, I.F.; Montague, S.C.; Houck, M.L.; Tran, H.T.; Garitaonandia, I.; Leonardo, T.R.; Wang, Y.C.; Charter, S.J.; Laurent, L.C.; Ryder, O.A.; et al. Induced pluripotent stem cells from highly endangered species. Nat. Methods 2011, 8, 829–831. [Google Scholar] [CrossRef]
- Comizzoli, P.; Holt, W.V. Recent advances and prospects in germplasm preservation of rare and endangered species. Adv. Exp. Med. Biol. 2014, 753, 331–356. [Google Scholar] [CrossRef] [PubMed]
- Mara, L.; Casu, S.; Carta, A.; Dattena, M. Cryobanking of farm animal gametes and embryos as a means of conserving livestock genetics. Anim. Reprod. Sci. 2013, 138, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.B.; Hermes, R.; Goeritz, F.; Appeltant, R.; Colleoni, S.; de Mori, B.; Diecke, S.; Drukker, M.; Galli, C.; Hayashi, K.; et al. The ART of bringing extinction to a freeze—History and future of species conservation, exemplified by rhinos. Theriogenology 2021, 169, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Dejosez, M.; Zwaka, T.P. Pluripotency and nuclear reprogramming. Annu. Rev. Biochem. 2012, 81, 737–765. [Google Scholar] [CrossRef] [PubMed]
- Pimm, S.L.; Alibhai, S.; Bergl, R.; Dehgan, A.; Giri, C.; Jewell, Z.; Joppa, L.; Kays, R.; Loarie, S. Emerging Technologies to Conserve Biodiversity. Trends Ecol. Evol. 2015, 30, 685–696. [Google Scholar] [CrossRef] [PubMed]
- Korody, M.L.; Ford, S.M.; Nguyen, T.D.; Pivaroff, C.G.; Valiente-Alandi, I.; Peterson, S.E.; Ryder, O.A.; Loring, J.F. Rewinding Extinction in the Northern White Rhinoceros: Genetically Diverse Induced Pluripotent Stem Cell Bank for Genetic Rescue. Stem Cells Dev. 2021, 30, 177–189. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Zywitza, V.; Naitou, Y.; Hamazaki, N.; Goeritz, F.; Hermes, R.; Holtze, S.; Lazzari, G.; Galli, C.; Stejskal, J.; et al. Robust induction of primordial germ cells of white rhinoceros on the brink of extinction. Sci. Adv. 2022, 8, eabp9683. [Google Scholar] [CrossRef]
- Katayama, M.; Hirayama, T.; Tani, T.; Nishimori, K.; Onuma, M.; Fukuda, T. Chick derived induced pluripotent stem cells by the poly-cistronic transposon with enhanced transcriptional activity. J. Cell Physiol. 2018, 233, 990–1004. [Google Scholar] [CrossRef]
- Kumar, D.; Talluri, T.R.; Selokar, N.L.; Hyder, I.; Kues, W.A. Perspectives of pluripotent stem cells in livestock. World J. Stem Cells 2021, 13, 1–29. [Google Scholar] [CrossRef]
- Verma, R.; Lee, Y.; Salamone, D.F. iPSC Technology: An Innovative Tool for Developing Clean Meat, Livestock, and Frozen Ark. Animals 2022, 12, 3187. [Google Scholar] [CrossRef]
- Stanton, M.M.; Tzatzalos, E.; Donne, M.; Kolundzic, N.; Helgason, I.; Ilic, D. Prospects for the Use of Induced Pluripotent Stem Cells in Animal Conservation and Environmental Protection. Stem Cells Transl. Med. 2019, 8, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Reiss, J.; Robertson, S.; Suzuki, M. Cell Sources for Cultivated Meat: Applications and Considerations throughout the Production Workflow. Int. J. Mol. Sci. 2021, 22, 7513. [Google Scholar] [CrossRef] [PubMed]
- Post, M.J.; Levenberg, S.; Kaplan, D.L.; Genovese, N.; Fu, J.A.; Bryant, C.J.; Negowetti, N.; Verzijden, K.; Moutsatsou, P. Scientific, sustainability and regulatory challenges of cultured meat. Nat. Food 2020, 1, 403–415. [Google Scholar] [CrossRef]
- Post, M.J. Cultured meat from stem cells: Challenges and prospects. Meat Sci. 2012, 92, 297–301. [Google Scholar] [CrossRef] [PubMed]
- Post, M.J. An alternative animal protein source: Cultured beef. Ann. Ny. Acad. Sci. 2014, 1328, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Genovese, N.J.; Domeier, T.L.; Telugu, B.P.; Roberts, R.M. Enhanced Development of Skeletal Myotubes from Porcine Induced Pluripotent Stem Cells. Sci. Rep. 2017, 7, 41833. [Google Scholar] [CrossRef] [PubMed]
- Devito, L.; Petrova, A.; Miere, C.; Codognotto, S.; Blakely, N.; Lovatt, A.; Ogilvie, C.; Khalaf, Y.; Ilic, D. Cost-effective master cell bank validation of multiple clinical-grade human pluripotent stem cell lines from a single donor. Stem Cells Transl. Med. 2014, 3, 1116–1124. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.; Hwang, Y.H.; Joo, S.T. Meat analog as future food: A review. J. Anim. Sci. Technol. 2020, 62, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Bhat, Z.F.; Kumar, S.; Fayaz, H. meat production: Challenges and benefits over conventional meat production. J. Integr. Agr. 2015, 14, 241–248. [Google Scholar] [CrossRef]
- Stephens, N.; Di Silvio, L.; Dunsford, I.; Ellis, M.; Glencross, A.; Sexton, A. Bringing cultured meat to market: Technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci. Technol. 2018, 78, 155–166. [Google Scholar] [CrossRef]
- Harding, J.; Roberts, R.M.; Mirochnitchenko, O. Large animal models for stem cell therapy. Stem Cell Res. Ther. 2013, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Dehghan, M.M.; Baghaban Eslaminejad, M.; Motallebizadeh, N.; Ashrafi Halan, J.; Tagiyar, L.; Soroori, S.; Nikmahzar, A.; Pedram, M.; Shahverdi, A.; Kazemi Mehrjerdi, H.; et al. Transplantation of Autologous Bone Marrow Mesenchymal Stem Cells with Platelet-Rich Plasma Accelerate Distraction Osteogenesis in A Canine Model. Cell J. 2015, 17, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Cong, X.; Zhang, S.M.; Ellis, M.W.; Luo, J. Large Animal Models for the Clinical Application of Human Induced Pluripotent Stem Cells. Stem Cells Dev. 2019, 28, 1288–1298. [Google Scholar] [CrossRef] [PubMed]
- Kehinde, E.O. They see a rat, we seek a cure for diseases: The current status of animal experimentation in medical practice. Med. Princ. Pract. 2013, 22 (Suppl. S1), 52–61. [Google Scholar] [CrossRef] [PubMed]
- Duranthon, V.; Beaujean, N.; Brunner, M.; Odening, K.E.; Santos, A.N.; Kacskovics, I.; Hiripi, L.; Weinstein, E.J.; Bosze, Z. On the emerging role of rabbit as human disease model and the instrumental role of novel transgenic tools. Transgenic Res. 2012, 21, 699–713. [Google Scholar] [CrossRef] [PubMed]
- Madeja, Z.E.; Pawlak, P.; Piliszek, A. Beyond the mouse: Non-rodent animal models for study of early mammalian development and biomedical research. Int. J. Dev. Biol. 2019, 63, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Plews, J.R.; Gu, M.; Longaker, M.T.; Wu, J.C. Large animal induced pluripotent stem cells as pre-clinical models for studying human disease. J. Cell Mol. Med. 2012, 16, 1196–1202. [Google Scholar] [CrossRef] [PubMed]
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabria, J.; Tibau, J. The pig as an animal model for human pathologies: A proteomics perspective. Proteom. Clin. Appl. 2014, 8, 715–731. [Google Scholar] [CrossRef]
- Klymiuk, N.; Seeliger, F.; Bohlooly, Y.M.; Blutke, A.; Rudmann, D.G.; Wolf, E. Tailored Pig Models for Preclinical Efficacy and Safety Testing of Targeted Therapies. Toxicol. Pathol. 2016, 44, 346–357. [Google Scholar] [CrossRef]
- Cebrian-Serrano, A.; Stout, T.; Dinnyes, A. Veterinary applications of induced pluripotent stem cells: Regenerative medicine and models for disease? Vet. J. 2013, 198, 34–42. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, W.; Liu, Y.; Fernandez de Castro, J.; Ezashi, T.; Telugu, B.P.; Roberts, R.M.; Kaplan, H.J.; Dean, D.C. Differentiation of induced pluripotent stem cells of swine into rod photoreceptors and their integration into the retina. Stem Cells 2011, 29, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Anand, T.; Kues, W.A. Clinical potential of human-induced pluripotent stem cells: Perspectives of induced pluripotent stem cells. Cell Biol. Toxicol. 2017, 33, 99–112. [Google Scholar] [CrossRef]
- Perleberg, C.; Kind, A.; Schnieke, A. Genetically engineered pigs as models for human disease. Dis. Model. Mech. 2018, 11, dmm030783. [Google Scholar] [CrossRef]
- Liao, Y.J.; Tang, P.C.; Chen, Y.H.; Chu, F.H.; Kang, T.C.; Chen, L.R.; Yang, J.R. Porcine induced pluripotent stem cell-derived osteoblast-like cells prevent glucocorticoid-induced bone loss in Lanyu pigs. PLoS ONE 2018, 13, e0202155. [Google Scholar] [CrossRef]
- Li, X.; Zhang, F.; Song, G.; Gu, W.; Chen, M.; Yang, B.; Li, D.; Wang, D.; Cao, K. Intramyocardial Injection of Pig Pluripotent Stem Cells Improves Left Ventricular Function and Perfusion: A Study in a Porcine Model of Acute Myocardial Infarction. PLoS ONE 2013, 8, e66688. [Google Scholar] [CrossRef]
- Malhi, P.S.; Adams, G.P.; Singh, J. Bovine model for the study of reproductive aging in women: Follicular, luteal, and endocrine characteristics. Biol. Reprod. 2005, 73, 45–53. [Google Scholar] [CrossRef]
- Cravero, D.; Martignani, E.; Miretti, S.; Accornero, P.; Pauciullo, A.; Sharma, R.; Donadeu, F.X.; Baratta, M. Generation of Induced Pluripotent Stem Cells from Bovine Epithelial Cells and Partial Redirection Toward a Mammary Phenotype In Vitro. Cell Reprogram 2015, 17, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Fortuna, P.R.J.; Bielefeldt-Ohmann, H.; Ovchinnikov, D.A.; Wolvetang, E.J.; Whitworth, D.J. Cortical Neurons Derived from Equine Induced Pluripotent Stem Cells Are Susceptible to Neurotropic Flavivirus Infection and Replication: An In Vitro Model for Equine Neuropathic Diseases. Stem Cells Dev. 2018, 27, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Bavin, E.P.; Smith, O.; Baird, A.E.; Smith, L.C.; Guest, D.J. Equine Induced Pluripotent Stem Cells have a Reduced Tendon Differentiation Capacity Compared to Embryonic Stem Cells. Front. Vet. Sci. 2015, 2, 55. [Google Scholar] [CrossRef]
- Amilon, K.R.; Cortes-Araya, Y.; Moore, B.; Lee, S.; Lillico, S.; Breton, A.; Esteves, C.L.; Donadeu, F.X. Generation of Functional Myocytes from Equine Induced Pluripotent Stem Cells. Cell Reprogram 2018, 20, 275–281. [Google Scholar] [CrossRef]
- Baird, A.; Dominguez Falcon, N.; Saeed, A.; Guest, D.J. Biocompatible Three-Dimensional Printed Thermoplastic Scaffold for Osteoblast Differentiation of Equine Induced Pluripotent Stem Cells. Tissue Eng. Part. C Methods 2019, 25, 253–261. [Google Scholar] [CrossRef]
- Sharma, R.; Livesey, M.R.; Wyllie, D.J.; Proudfoot, C.; Whitelaw, C.B.; Hay, D.C.; Donadeu, F.X. Generation of functional neurons from feeder-free, keratinocyte-derived equine induced pluripotent stem cells. Stem Cells Dev. 2014, 23, 1524–1534. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, C.; Therrien, J.; Lemire, P.; Segura, M.; Smith, L.C.; Theoret, C.L. Differentiation of equine induced pluripotent stem cells into a keratinocyte lineage. Equine Vet. J. 2016, 48, 338–345. [Google Scholar] [CrossRef]
- Scarfone, R.A.; Pena, S.M.; Russell, K.A.; Betts, D.H.; Koch, T.G. The use of induced pluripotent stem cells in domestic animals: A narrative review. Bmc Vet. Res. 2020, 16, 477. [Google Scholar] [CrossRef] [PubMed]
- Whitworth, D.J.; Frith, J.E.; Frith, T.J.; Ovchinnikov, D.A.; Cooper-White, J.J.; Wolvetang, E.J. Derivation of mesenchymal stromal cells from canine induced pluripotent stem cells by inhibition of the TGFbeta/activin signaling pathway. Stem Cells Dev. 2014, 23, 3021–3033. [Google Scholar] [CrossRef] [PubMed]
- Susta, L.; He, Y.; Hutcheson, J.M.; Lu, Y.; West, F.D.; Stice, S.L.; Yu, P.; Abdo, Z.; Afonso, C.L. Derivation of chicken induced pluripotent stem cells tolerant to Newcastle disease virus-induced lysis through multiple rounds of infection. Virol. J. 2016, 13, 205. [Google Scholar] [CrossRef]
- Sutton, T.C. The Pandemic Threat of Emerging H5 and H7 Avian Influenza Viruses. Viruses 2018, 10, 461. [Google Scholar] [CrossRef] [PubMed]
- Liou, J.F.; Wu, W.R.; Chen, L.R.; Shiue, Y.L. Establishment of an induced pluripotent cell line from Taiwan black silkie chick embryonic fibroblasts for replication-incompetent virus production. Sci. Rep. 2019, 9, 15745. [Google Scholar] [CrossRef]
- Lee, E.M.; Kim, A.Y.; Lee, E.J.; Park, J.K.; Park, S.I.; Cho, S.G.; Kim, H.K.; Kim, S.Y.; Jeong, K.S. Generation of Equine-Induced Pluripotent Stem Cells and Analysis of Their Therapeutic Potential for Muscle Injuries. Cell Transpl. 2016, 25, 2003–2016. [Google Scholar] [CrossRef]
- Chung, M.J.; Park, S.; Son, J.Y.; Lee, J.Y.; Yun, H.H.; Lee, E.J.; Lee, E.M.; Cho, G.J.; Lee, S.; Park, H.S.; et al. Differentiation of equine induced pluripotent stem cells into mesenchymal lineage for therapeutic use. Cell Cycle 2019, 18, 2954–2971. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Gong, W.; Yao, Z.; Jin, K.; Niu, Y.; Li, B.; Zuo, Q. Mechanisms of Embryonic Stem Cell Pluripotency Maintenance and Their Application in Livestock and Poultry Breeding. Animals 2024, 14, 1742. https://doi.org/10.3390/ani14121742
Wang Z, Gong W, Yao Z, Jin K, Niu Y, Li B, Zuo Q. Mechanisms of Embryonic Stem Cell Pluripotency Maintenance and Their Application in Livestock and Poultry Breeding. Animals. 2024; 14(12):1742. https://doi.org/10.3390/ani14121742
Chicago/Turabian StyleWang, Ziyu, Wei Gong, Zeling Yao, Kai Jin, Yingjie Niu, Bichun Li, and Qisheng Zuo. 2024. "Mechanisms of Embryonic Stem Cell Pluripotency Maintenance and Their Application in Livestock and Poultry Breeding" Animals 14, no. 12: 1742. https://doi.org/10.3390/ani14121742
APA StyleWang, Z., Gong, W., Yao, Z., Jin, K., Niu, Y., Li, B., & Zuo, Q. (2024). Mechanisms of Embryonic Stem Cell Pluripotency Maintenance and Their Application in Livestock and Poultry Breeding. Animals, 14(12), 1742. https://doi.org/10.3390/ani14121742





