The Association between Broiler Litter Microbiota and the Supplementation of Bacillus Probiotics in a Leaky Gut Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction and Sequencing
2.3. Litter Moisture Content and pH
2.4. Bioinformatics
3. Results
3.1. Animal Performance
3.2. Litter Microbiota Richness and Diversity
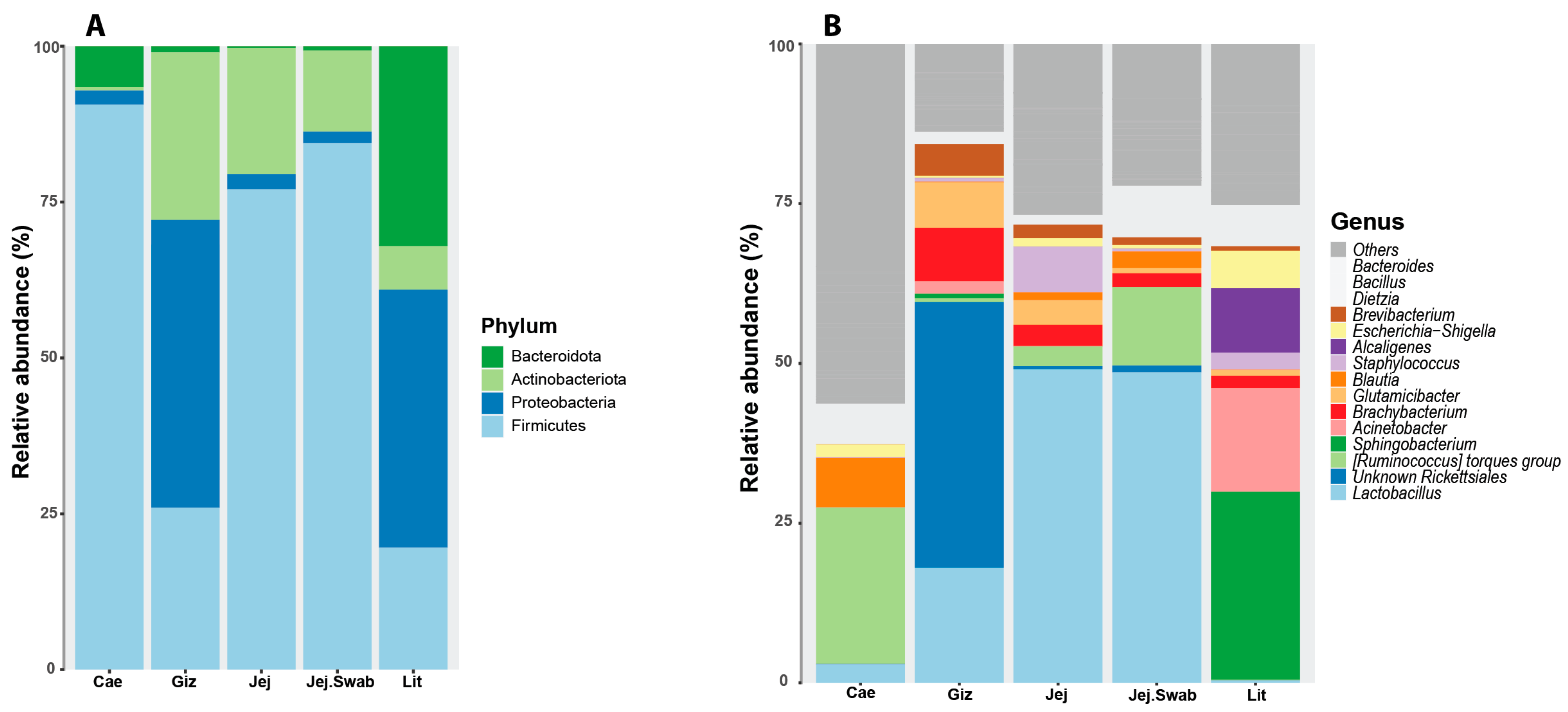
3.3. Litter Microbial Community
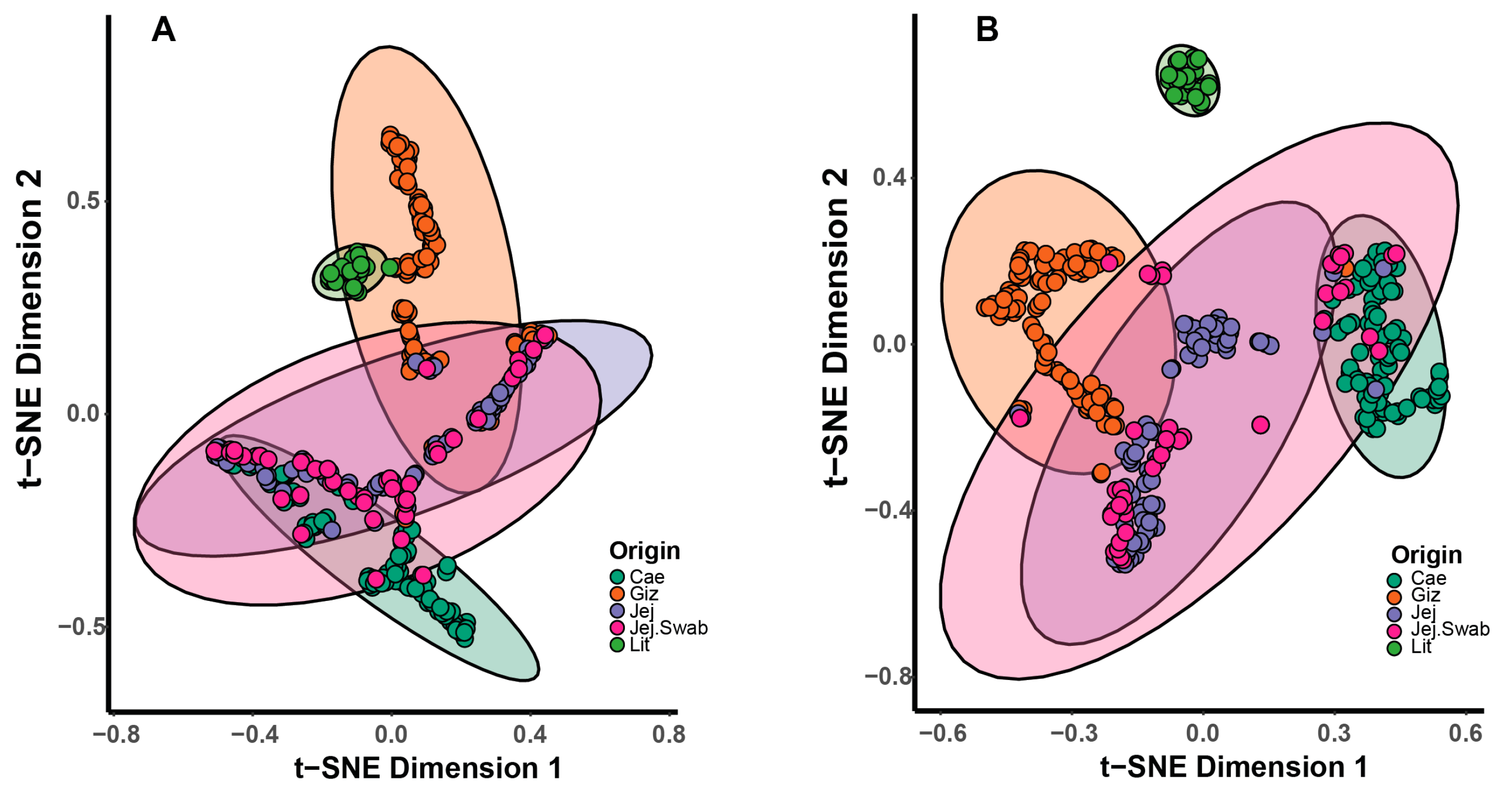
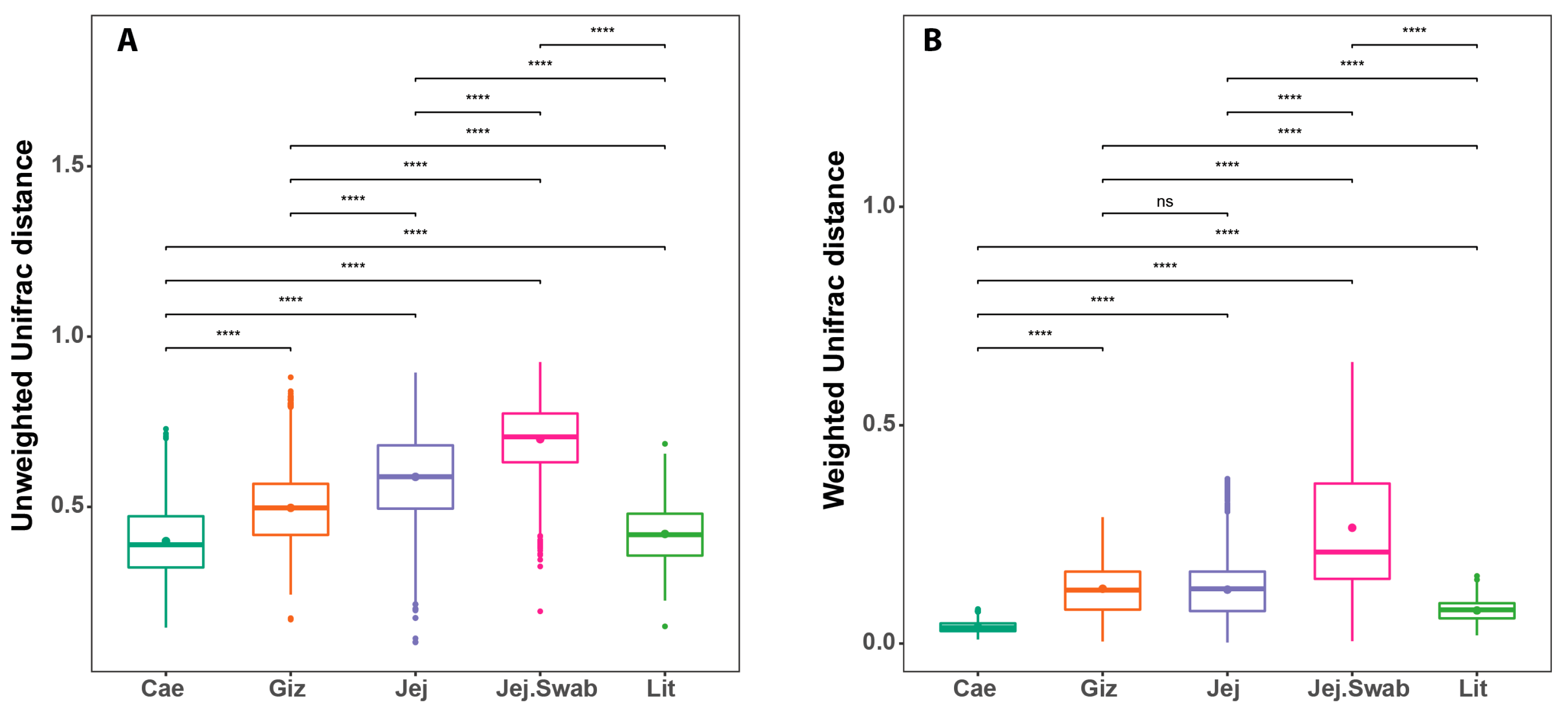
3.4. Beta Diversity
3.5. Effects of Treatment on Litter Microbial Taxa
3.6. Litter Quality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pan, D.; Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes 2014, 5, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Rychlik, I. Composition and Function of Chicken Gut Microbiota. Animals 2020, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.J.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Aruwa, C.E.; Pillay, C.; Nyaga, M.M.; Sabiu, S. Poultry gut health-microbiome functions, environmental impacts, microbiome engineering and advancements in characterization technologies. J. Anim. Sci. Biotechnol. 2021, 12, 119. [Google Scholar] [CrossRef] [PubMed]
- Wise, M.G.; Siragusa, G.R. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 2007, 102, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Schokker, D.; Hoekman, A.J.W.; Smits, M.A.; Rebel, J.M.J. Gene expression patterns associated with chicken jejunal development. Dev. Comp. Immunol. 2009, 33, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Bindari, Y.R.; Moore, R.J.; Van, T.T.H.; Hilliar, M.; Wu, S.B.; Walkden-Brown, S.W.; Gerber, P.F. Microbial communities of poultry house dust, excreta and litter are partially representative of microbiota of chicken caecum and ileum. PLoS ONE 2021, 16, e0255633. [Google Scholar] [CrossRef] [PubMed]
- Bajagai, Y.S.; Van, T.T.H.; Joat, N.; Chousalkar, K.; Moore, R.J.; Stanley, D. Layer chicken microbiota: A comprehensive analysis of spatial and temporal dynamics across all major gut sections. J. Anim. Sci. Biotechnol. 2024, 15, 20. [Google Scholar] [CrossRef]
- Ahaduzzaman, M.; Groves, P.J.; Sharpe, S.M.; Williamson, S.L.; Gao, Y.K.; Nguyen, T.V.; Gerber, P.F.; Walkden-Brown, S.W. A practical method for assessing infectious laryngotracheitis vaccine take in broilers following mass administration in water: Spatial and temporal variation in viral genome content of poultry dust after vaccination. Vet. Microbiol. 2020, 241, 108545. [Google Scholar] [CrossRef]
- Assen, A.M.; Stillman, M.; Alfirevich, S.; Gerber, P.F.; Groves, P.J.; Walkden-Brown, S.W. Assessment of A20 infectious laryngotracheitis vaccine take in meat chickens using swab and dust samples following mass vaccination in drinking water. Vet. Microbiol. 2020, 251, 108903. [Google Scholar] [CrossRef]
- Ahaduzzaman, M.; Keerqin, C.; Kumar, A.; Musigwa, S.; Morgan, N.; Kheravii, S.K.; Sharpe, S.; Williamson, S.; Wu, S.-B.; Walkden-Brown, S.W. Detection and quantification of Clostridium perfringens and Eimeria spp. in poultry dust using real-time PCR under experimental and field conditions. Avian Dis. 2021, 65, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Liebhart, D.; Bilic, I.; Grafl, B.; Hess, C.; Hess, M. Diagnosing Infectious Diseases in Poultry Requires a Holistic Approach: A Review. Poultry 2023, 2, 252–280. [Google Scholar] [CrossRef]
- Diaz Carrasco, J.M.; Casanova, N.A.; Fernández Miyakawa, M.E. Microbiota, Gut Health and Chicken Productivity: What Is the Connection? Microorganisms 2019, 7, 374. [Google Scholar] [CrossRef] [PubMed]
- Mulchandani, R.; Wang, Y.; Gilbert, M.; Van Boeckel, T.P. Global trends in antimicrobial use in food-producing animals: 2020 to 2030. PLoS Glob. Public Health 2023, 3, e0001305. [Google Scholar] [CrossRef] [PubMed]
- Kogut, M.H.; Genovese, K.J.; Swaggerty, C.L.; He, H.; Broom, L. Inflammatory phenotypes in the intestine of poultry: Not all inflammation is created equal. Poult. Sci. 2018, 97, 2339–2346. [Google Scholar] [CrossRef] [PubMed]
- Feye, K.M.; Baxter, M.F.A.; Tellez-Isaias, G.; Kogut, M.H.; Ricke, S.C. Influential factors on the composition of the conventionally raised broiler gastrointestinal microbiomes. Poult. Sci. 2020, 99, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Cockerill, S.A.; Gerber, P.F.; Walkden-Brown, S.W.; Dunlop, M.W. Suitability of litter amendments for the Australian chicken meat industry. Anim. Prod. Sci. 2020, 60, 1469–1481. [Google Scholar] [CrossRef]
- Cressman, M.D.; Yu, Z.; Nelson, M.C.; Moeller, S.J.; Lilburn, M.S.; Zerby, H.N. Interrelations between the microbiotas in the litter and in the intestines of commercial broiler chickens. Appl. Environ. Microbiol. 2010, 76, 6572–6582. [Google Scholar] [CrossRef]
- Wei, S.; Morrison, M.; Yu, Z. Bacterial census of poultry intestinal microbiome. Poult. Sci. 2013, 92, 671–683. [Google Scholar] [CrossRef]
- Stanley, V.; Gray, C.; Daley, M.; Krueger, W.; Sefton, A. An alternative to antibiotic-based drugs in feed for enhancing performance of broilers grown on Eimeria spp.-infected litter. Poult. Sci. 2004, 83, 39–44. [Google Scholar] [CrossRef]
- Cambra-López, M.; Hermosilla, T.; Lai, H.T.; Montero, M.; Aarnink, A.J.; Ogink, N.W. Source identification and quantification of particulate matter emitted from livestock houses. In Proceedings of the International Symposium on Air Quality and Manure Management for Agriculture Conference Proceedings, Dallas, TX, USA, 13–16 September 2010; p. 41. [Google Scholar]
- O’Brien, K.M.; Chimenti, M.S.; Farnell, M.; Tabler, T.; Bair, T.; Bray, J.L.; Nonnenmann, M.W. High throughput genomic sequencing of bioaerosols in broiler chicken production facilities. Microb. Biotechnol. 2016, 9, 782–791. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Sun, C.; Zheng, J.; Wen, C.; Ji, C.; Zhang, D.; Chen, Y.; Hou, Z.; Yang, N. Efficacy of Fecal Sampling as a Gut Proxy in the Study of Chicken Gut Microbiota. Front. Microbiol. 2019, 10, 2126. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Kfzumar, S.; Oakley, B.; Kim, W.K. Chicken Gut Microbiota: Importance and Detection Technology. Front. Vet. Sci. 2018, 5, 254. [Google Scholar] [CrossRef] [PubMed]
- Dunlop, M.W.; McAuley, J.; Blackall, P.J.; Stuetz, R.M. Water activity of poultry litter: Relationship to moisture content during a grow-out. J. Environ. Manag. 2016, 172, 201–206. [Google Scholar] [CrossRef]
- Krysiak, K.; Konkol, D.; Korczyński, M. Overview of the Use of Probiotics in Poultry Production. Animals 2021, 11, 1620. [Google Scholar] [CrossRef] [PubMed]
- Ogbuewu, I.P.; Mabelebele, M.; Sebola, N.A.; Mbajiorgu, C. Bacillus Probiotics as Alternatives to In-feed Antibiotics and Its Influence on Growth, Serum Chemistry, Antioxidant Status, Intestinal Histomorphology, and Lesion Scores in Disease-Challenged Broiler Chickens. Front. Vet. Sci. 2022, 9, 876725. [Google Scholar] [CrossRef] [PubMed]
- Attia, Y.A.; Basiouni, S.; Abdulsalam, N.M.; Bovera, F.; Aboshok, A.A.; Shehata, A.A.; Hafez, H.M. Alternative to antibiotic growth promoters: Beneficial effects of Saccharomyces cerevisiae and/or Lactobacillus acidophilus supplementation on the growth performance and sustainability of broilers’ production. Front. Vet. Sci. 2023, 10, 1259426. [Google Scholar] [CrossRef]
- Costa, F.G.P.; Goulart, C.; Figueiredo, D.; Oliveira, C.; Silva, J. Economic and environmental impact of using exogenous enzymes on poultry feeding. Int. J. Poult. Sci. 2008, 7, 311–314. [Google Scholar]
- Vicuña, E.A.; Kuttappan, V.A.; Galarza-Seeber, R.; Latorre, J.D.; Faulkner, O.B.; Hargis, B.M.; Tellez, G.; Bielke, L.R. Effect of dexamethasone in feed on intestinal permeability, differential white blood cell counts, and immune organs in broiler chicks. Poult. Sci. 2015, 94, 2075–2080. [Google Scholar] [CrossRef]
- Horyanto, D.; Bajagai, Y.S.; Kayal, A.; von Hellens, J.; Chen, X.; Van, T.T.H.; Radovanović, A.; Stanley, D. Bacillus amyloliquefaciens Probiotics Mix Supplementation in a Broiler Leaky Gut Model. Microorganisms 2024, 12, 419. [Google Scholar] [CrossRef]
- Yu, Z.; Morrison, M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques 2004, 36, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Elias, D.M.O.; Robinson, S.; Both, S.; Goodall, T.; Majalap-Lee, N.; Ostle, N.J.; McNamara, N.P. Soil Microbial Community and Litter Quality Controls on Decomposition Across a Tropical Forest Disturbance Gradient. Front. For. Glob. Chang. 2020, 3, 81. [Google Scholar] [CrossRef]
- Martin, M. CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Smith, S. phylosmith: An R-package for reproducible and efficient microbiome analysis with phyloseq-objects. J. Open Source Softw. 2019, 4, 1442. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tellez, G.; Richards, J.D.; Escobar, J. Identification of Potential Biomarkers for Gut Barrier Failure in Broiler Chickens. Front. Vet. Sci. 2015, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Awad, W.A.; Hess, C.; Hess, M. Enteric Pathogens and Their Toxin-Induced Disruption of the Intestinal Barrier through Alteration of Tight Junctions in Chickens. Toxins 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Cardoso Dal Pont, G.; Farnell, M.; Farnell, Y.; Kogut, M.H. Dietary Factors as Triggers of Low-Grade Chronic Intestinal Inflammation in Poultry. Microorganisms 2020, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Duckworth, C.A.; Burkitt, M.D.; Watson, A.J.; Campbell, B.J.; Pritchard, D.M. Epithelial cell shedding and barrier function: A matter of life and death at the small intestinal villus tip. Vet. Pathol. 2015, 52, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Hanhimäki, E.; Watts, P.C.; Koskela, E.; Koteja, P.; Mappes, T.; Hämäläinen, A.M. Evolved high aerobic capacity has context-specific effects on gut microbiota. Front. Ecol. Evol. 2022, 10, 934164. [Google Scholar] [CrossRef]
- Maki, J.J.; Klima, C.L.; Sylte, M.J.; Looft, T. The Microbial Pecking Order: Utilization of Intestinal Microbiota for Poultry Health. Microorganisms 2019, 7, 376. [Google Scholar] [CrossRef] [PubMed]
- Fancher, C.A.; Zhang, L.; Kiess, A.S.; Adhikari, P.A.; Dinh, T.T.N.; Sukumaran, A.T. Avian Pathogenic Escherichia coli and Clostridium perfringens: Challenges in No Antibiotics Ever Broiler Production and Potential Solutions. Microorganisms 2020, 8, 1533. [Google Scholar] [CrossRef]
- Choct, M.; Hughes, R.J.; Wang, J.; Bedford, M.R.; Morgan, A.J.; Annison, G. Increased small intestinal fermentation is partly responsible for the anti-nutritive activity of non-starch polysaccharides in chickens. Br. Poult. Sci. 1996, 37, 609–621. [Google Scholar] [CrossRef]
- Teider, P.I.; Ribeiro, J.C.; Ossugui, E.H.; Tamanini, R.; Ribeiro, J.; Santos, G.A.; Alfieri, A.A.; Beloti, V. Pseudomonas spp. and other psychrotrophic microorganisms in inspected and non-inspected Brazilian Minas Frescal cheese: Proteolytic, lipolytic and AprX production potential. Pesqui. Veterinária Bras. 2019, 39, 807–815. [Google Scholar] [CrossRef]
- Yehia, N.; Salem, H.M.; Mahmmod, Y.; Said, D.; Samir, M.; Mawgod, S.A.; Sorour, H.K.; AbdelRahman, M.A.A.; Selim, S.; Saad, A.M.; et al. Common viral and bacterial avian respiratory infections: An updated review. Poult. Sci. 2023, 102, 102553. [Google Scholar] [CrossRef] [PubMed]
- Campos, P.M.; Darwish, N.; Shao, J.; Proszkowiec-Weglarz, M. Research Note: Choice of microbiota database affects data analysis and interpretation in chicken cecal microbiota. Poult. Sci. 2022, 101, 101971. [Google Scholar] [CrossRef] [PubMed]
- Temple, L.M.; Weiss, A.A.; Walker, K.E.; Barnes, H.J.; Christensen, V.L.; Miyamoto, D.M.; Shelton, C.B.; Orndorff, P.E. Bordetella avium virulence measured in vivo and in vitro. Infect. Immun. 1998, 66, 5244–5251. [Google Scholar] [CrossRef] [PubMed]
- Van Immerseel, F.; De Buck, J.; Pasmans, F.; Huyghebaert, G.; Haesebrouck, F.; Ducatelle, R. Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 2004, 33, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.; Batool, A.; Arif, S. Healthy Cattle Microbiome and Dysbiosis in Diseased Phenotypes. Ruminants 2022, 2, 134–156. [Google Scholar] [CrossRef]
- Mally, M.; Shin, H.; Paroz, C.; Landmann, R.; Cornelis, G.R. Capnocytophaga canimorsus: A human pathogen feeding at the surface of epithelial cells and phagocytes. PLoS Pathog. 2008, 4, e1000164. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chai, B. Fate of Antibiotic Resistance Genes and Changes in Bacterial Community with Increasing Breeding Scale of Layer Manure. Front. Microbiol. 2022, 13, 857046. [Google Scholar] [CrossRef]
- Abdel-Kafy, E.-S.M.; Youssef, S.F.; Magdy, M.; Ghoneim, S.S.; Abdelatif, H.A.; Deif-Allah, R.A.; Abdel-Ghafar, Y.Z.; Shabaan, H.M.A.; Liu, H.; Elokil, A. Gut Microbiota, Intestinal Morphometric Characteristics, and Gene Expression in Relation to the Growth Performance of Chickens. Animals 2022, 12, 3474. [Google Scholar] [CrossRef]
- Mon, K.K.; Zhu, Y.; Chanthavixay, G.; Kern, C.; Zhou, H. Integrative analysis of gut microbiome and metabolites revealed novel mechanisms of intestinal Salmonella carriage in chicken. Sci. Rep. 2020, 10, 4809. [Google Scholar] [CrossRef]
- Wilkinson, T.J.; Cowan, A.A.; Vallin, H.E.; Onime, L.A.; Oyama, L.B.; Cameron, S.J.; Gonot, C.; Moorby, J.M.; Waddams, K.; Theobald, V.J.; et al. Characterization of the Microbiome along the Gastrointestinal Tract of Growing Turkeys. Front. Microbiol. 2017, 8, 1089. [Google Scholar] [CrossRef]
- Montecillo, J.A.V. Phylogenomics and comparative genomic analyses support the creation of the novel family Ignatzschineriaceae fam. nov. comprising the genera Ignatzschineria and Wohlfahrtiimonas within the order Cardiobacteriales. Res. Microbiol. 2023, 174, 103988. [Google Scholar] [CrossRef] [PubMed]
- Kopf, A.; Bunk, B.; Riedel, T.; Schröttner, P. The zoonotic pathogen Wohlfahrtiimonas chitiniclastica—Current findings from a clinical and genomic perspective. BMC Microbiol. 2024, 24, 3. [Google Scholar] [CrossRef] [PubMed]
- Maki, J.J.; Bobeck, E.A.; Sylte, M.J.; Looft, T. Eggshell and environmental bacteria contribute to the intestinal microbiota of growing chickens. J. Anim. Sci. Biotechnol. 2020, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Y.; Cottrell, J.J.; Suleria, H.A.R.; Dunshea, F.R. Gut Microbiota-Polyphenol Interactions in Chicken: A Review. Animals 2020, 10, 1391. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Pena, P.; Cervantes, R.; Dias, M.; Viegas, C. Microbial Contamination of Bedding Material: One Health in Poultry Production. Int. J. Environ. Res. Public Health 2022, 19, 16508. [Google Scholar] [CrossRef] [PubMed]
- Chinivasagam, H.N.; Tran, T.; Blackall, P.J. Impact of the Australian litter re-use practice on Salmonella in the broiler farming environment. Food Res. Int. 2012, 45, 891–896. [Google Scholar] [CrossRef]
- Abd El-Hamid, M.I.; Awad, N.F.S.; Hashem, Y.M.; Abdel-Rahman, M.A.; Abdelaziz, A.M.; Mohammed, I.A.A.; Abo-Shama, U.H. In vitro evaluation of various antimicrobials against field mycoplasma gallisepticum and mycoplasma synoviae isolates in Egypt. Poult. Sci. 2019, 98, 6281–6288. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.K.; Pajarillo, E.A.B.; Chae, J.P.; Kim, I.H.; Kang, D.K. Protective effects of Bacillus subtilis against Salmonella infection in the microbiome of Hy-Line Brown layers. Asian-Australas J. Anim. Sci. 2017, 30, 1332–1339. [Google Scholar] [CrossRef]
- Knap, I.; Kehlet, A.B.; Bennedsen, M.; Mathis, G.F.; Hofacre, C.L.; Lumpkins, B.S.; Jensen, M.M.; Raun, M.; Lay, A. Bacillus subtilis (DSM17299) significantly reduces Salmonella in broilers. Poult. Sci. 2011, 90, 1690–1694. [Google Scholar] [CrossRef]
- La Ragione, R.M.; Woodward, M.J. Competitive exclusion by Bacillus subtilis spores of Salmonella enterica serotype Enteritidis and Clostridium perfringens in young chickens. Vet. Microbiol. 2003, 94, 245–256. [Google Scholar] [CrossRef]



| Probiotic Strains | Specification |
|---|---|
| Bacillus amyloliquefaciens (BPR-11) | 2 × 108 CFU/g |
| Bacillus amyloliquefaciens (BPR-16) | 2 × 108 CFU/g |
| Bacillus amyloliquefaciens (BPR-17) | 2 × 108 CFU/g |
| Groups | R2 | p-Value | Distance |
|---|---|---|---|
| DEX | 0.111564 | 0.001 * | Unweighted UniFrac |
| Probiotic | 0.032266 | 0.710 | Unweighted UniFrac |
| Treatments | 0.172054 | 0.010 * | Unweighted UniFrac |
| DEX | 0.119781 | 0.022 * | Weighted UniFrac |
| Probiotic | 0.010500 | 0.933 | Weighted UniFrac |
| Treatments | 0.140897 | 0.287 | Weighted UniFrac |
| Groups | R2 | p (>F) | Distance |
|---|---|---|---|
| C vs. CD | 0.153235 | 0.007 * | Unweighted UniFrac |
| C vs. P | 0.079020 | 0.563 | Unweighted UniFrac |
| C vs. PD | 0.123330 | 0.069 | Unweighted UniFrac |
| CD vs. P | 0.168461 | 0.001 * | Unweighted UniFrac |
| CD vs. PD | 0.056801 | 0.849 | Unweighted UniFrac |
| P vs. PD | 0.137487 | 0.021 * | Unweighted UniFrac |
| C vs. CD | 0.118515 | 0.257 | Weighted UniFrac |
| C vs. P | 0.021170 | 0.943 | Weighted UniFrac |
| C vs. PD | 0.175991 | 0.078 | Weighted UniFrac |
| CD vs. P | 0.091673 | 0.289 | Weighted UniFrac |
| CD vs. PD | 0.026885 | 0.878 | Weighted UniFrac |
| P vs. PD | 0.142875 | 0.085 | Weighted UniFrac |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horyanto, D.; Bajagai, Y.S.; von Hellens, J.; Chen, X.; Thi Thu Hao, V.; Dunlop, M.W.; Stanley, D. The Association between Broiler Litter Microbiota and the Supplementation of Bacillus Probiotics in a Leaky Gut Model. Animals 2024, 14, 1758. https://doi.org/10.3390/ani14121758
Horyanto D, Bajagai YS, von Hellens J, Chen X, Thi Thu Hao V, Dunlop MW, Stanley D. The Association between Broiler Litter Microbiota and the Supplementation of Bacillus Probiotics in a Leaky Gut Model. Animals. 2024; 14(12):1758. https://doi.org/10.3390/ani14121758
Chicago/Turabian StyleHoryanto, Darwin, Yadav S. Bajagai, Juhani von Hellens, Xiaojing Chen, Van Thi Thu Hao, Mark W. Dunlop, and Dragana Stanley. 2024. "The Association between Broiler Litter Microbiota and the Supplementation of Bacillus Probiotics in a Leaky Gut Model" Animals 14, no. 12: 1758. https://doi.org/10.3390/ani14121758
APA StyleHoryanto, D., Bajagai, Y. S., von Hellens, J., Chen, X., Thi Thu Hao, V., Dunlop, M. W., & Stanley, D. (2024). The Association between Broiler Litter Microbiota and the Supplementation of Bacillus Probiotics in a Leaky Gut Model. Animals, 14(12), 1758. https://doi.org/10.3390/ani14121758








