An In Vitro System Mimics the Intestinal Microbiota of Striped Beakfish (Oplegnathus fasciatus) and Inhibits Vibrio alginolyticus by Limosilactobacillus reuteri-Derived Extracellular Vesicles
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Samples Preparation
2.3. Culture of Intestinal Bacteria and Microbiota Sequencing
2.4. The Effects of L. reuteri-Derived EVs against Vibrio Alginolyticus
2.5. Prediction of Small RNA Obtained from L. reuteri-Derived EVs
2.6. Statistical Analysis
3. Results
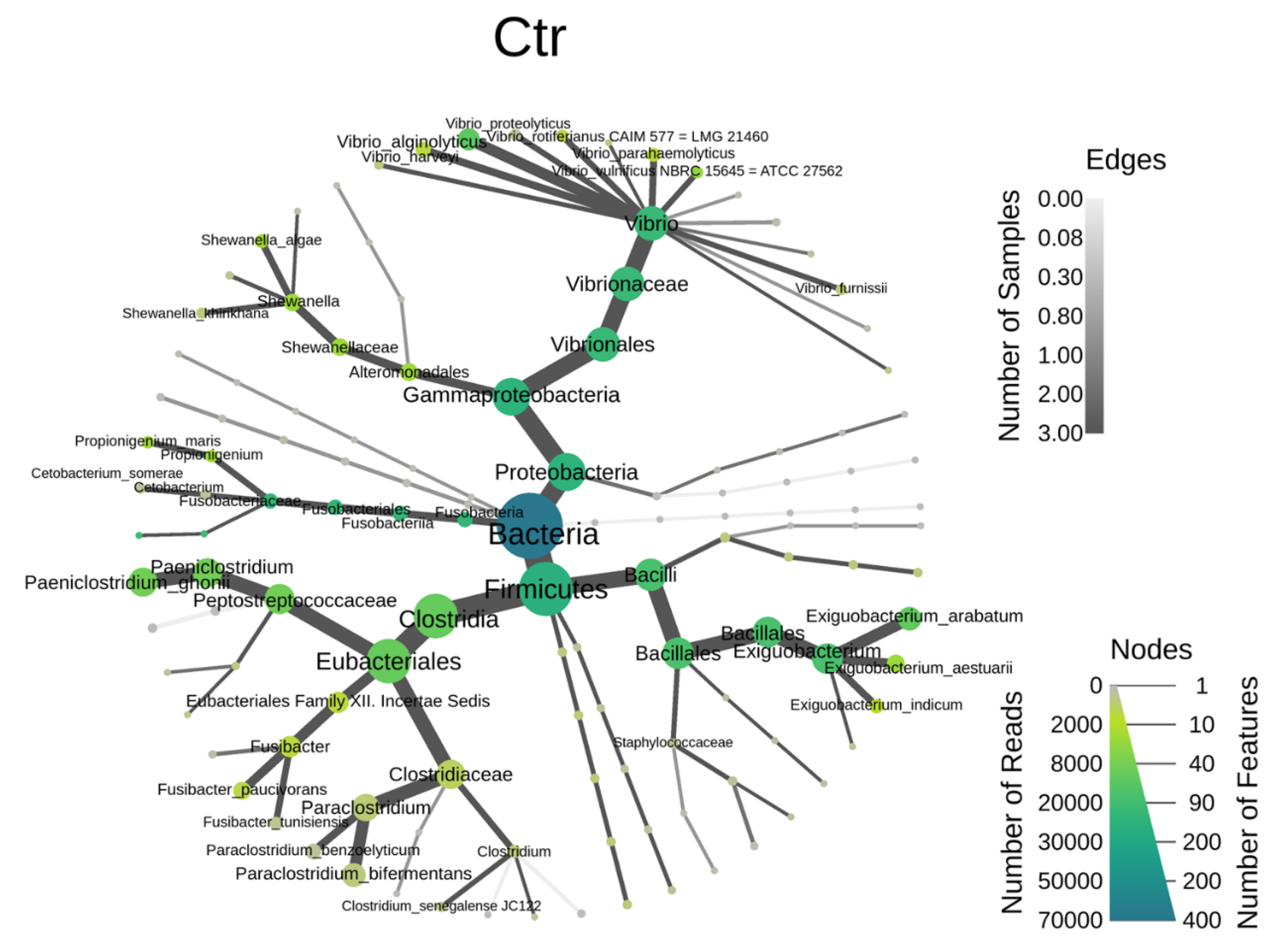
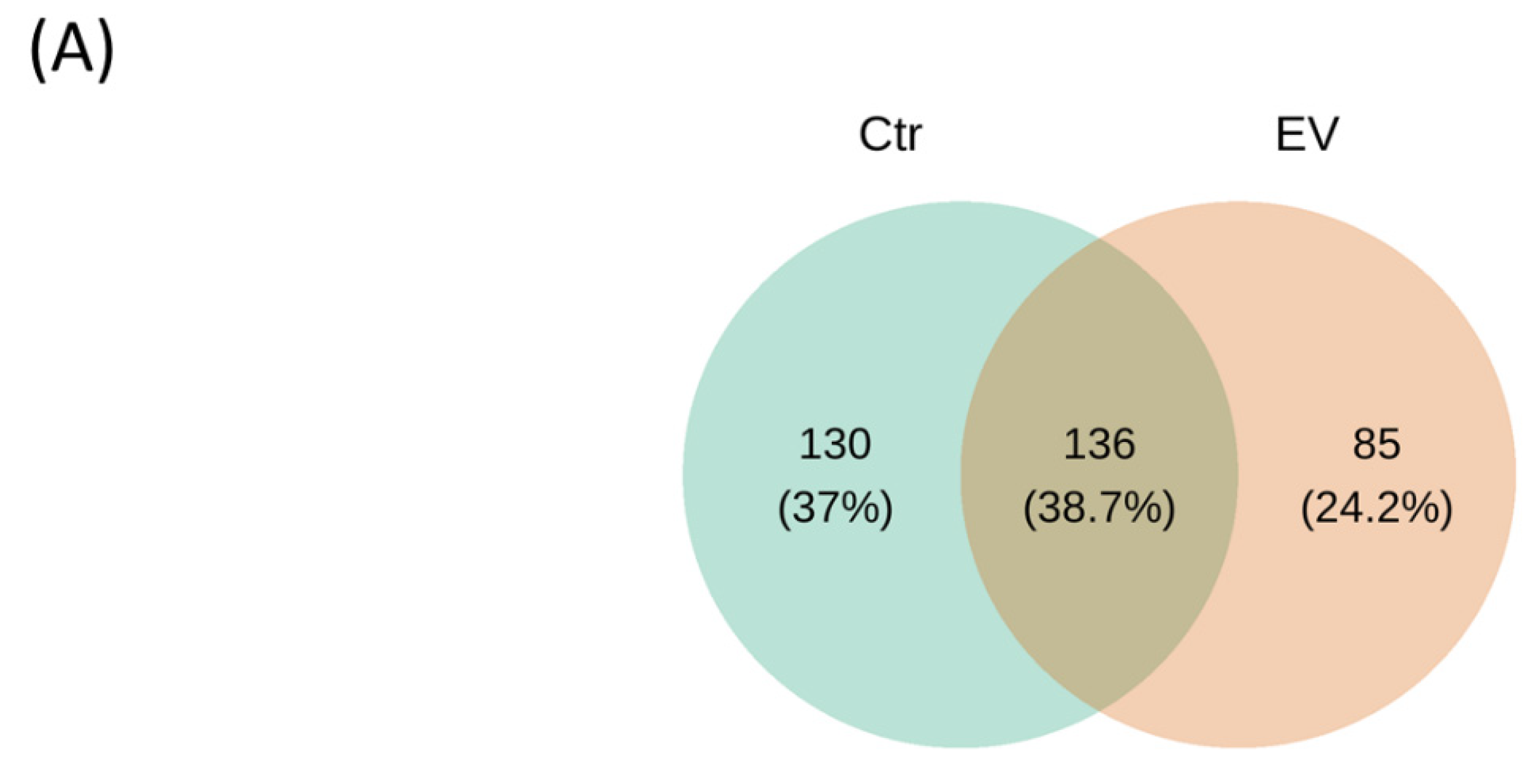
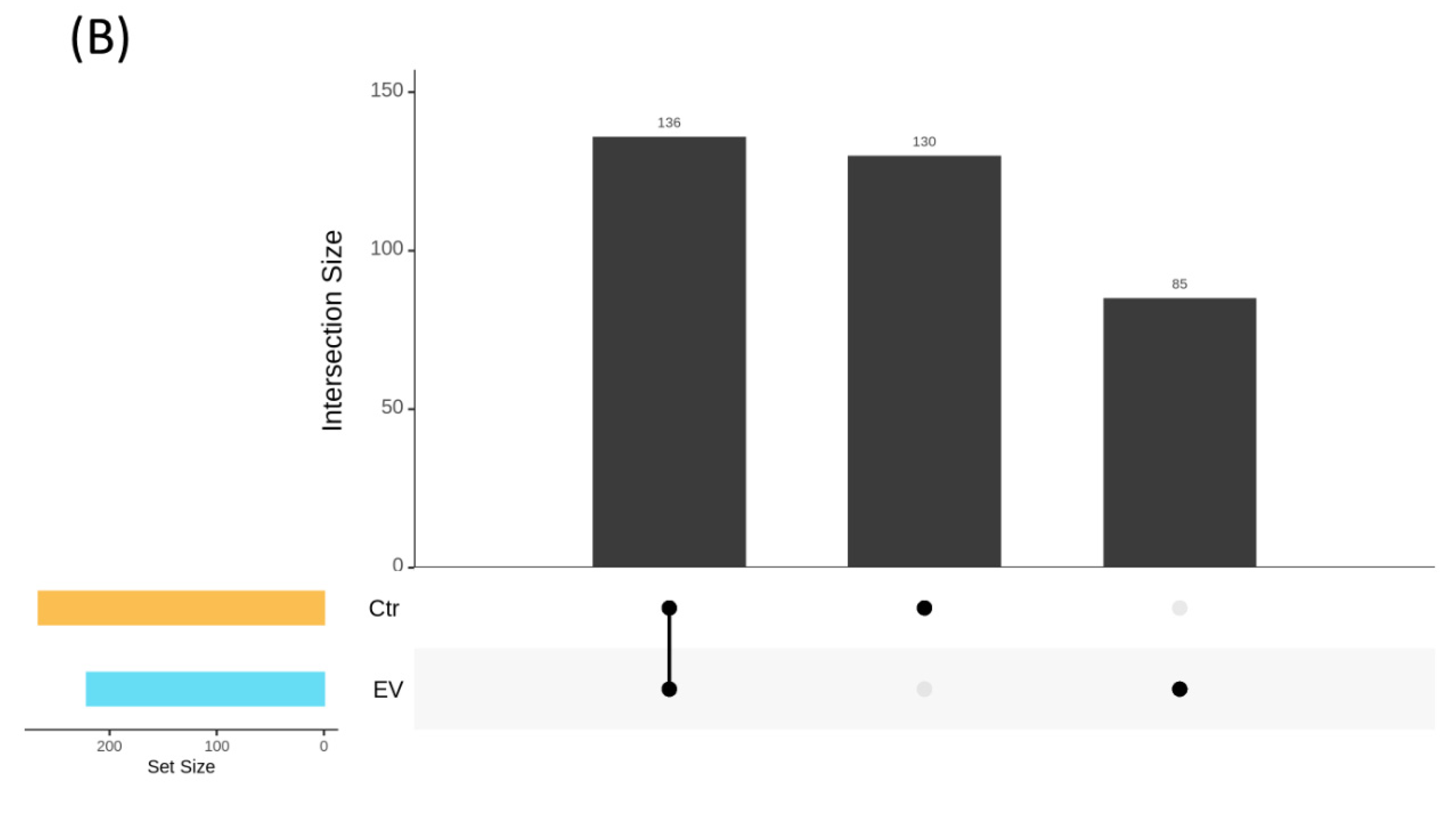
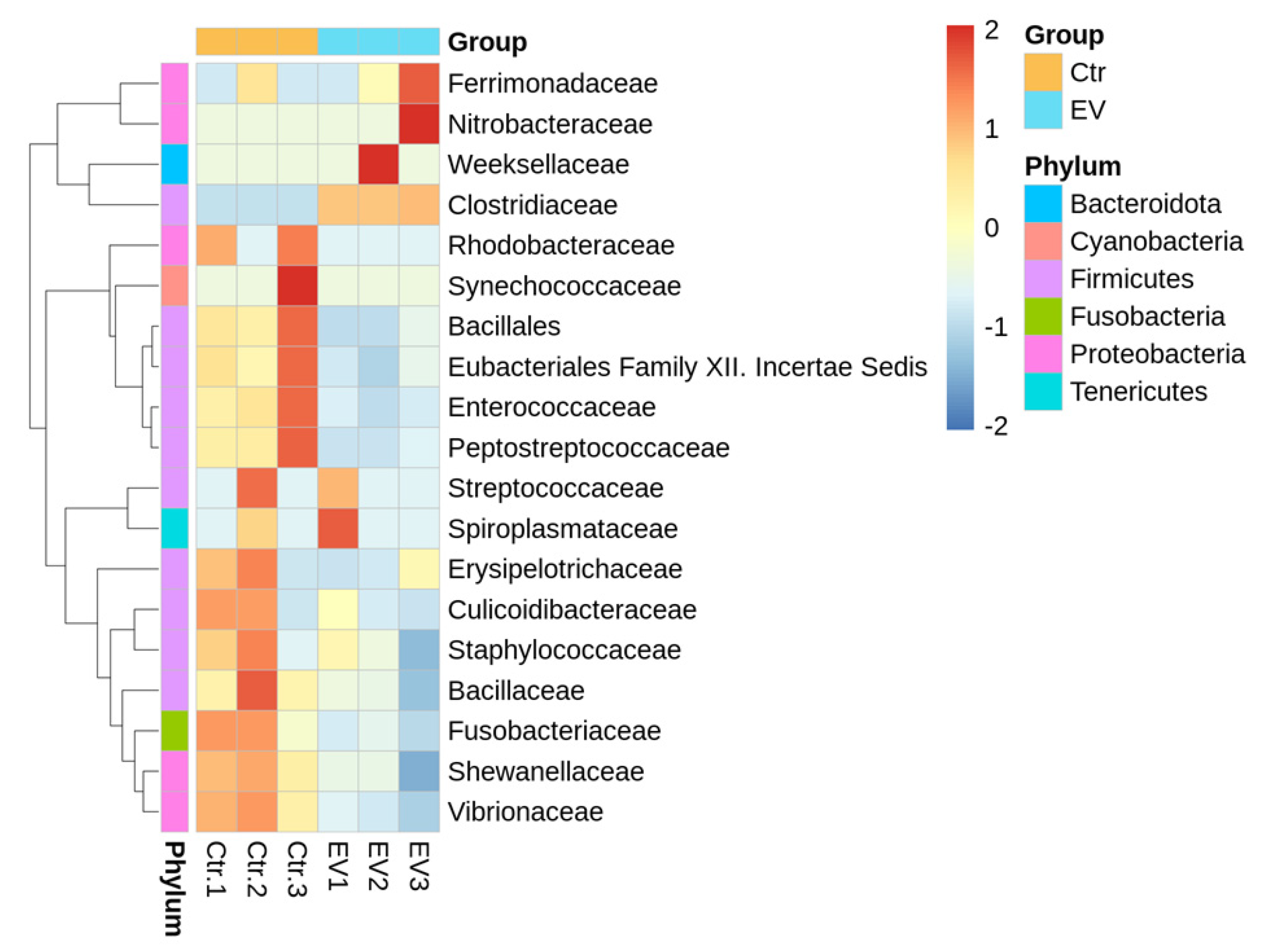
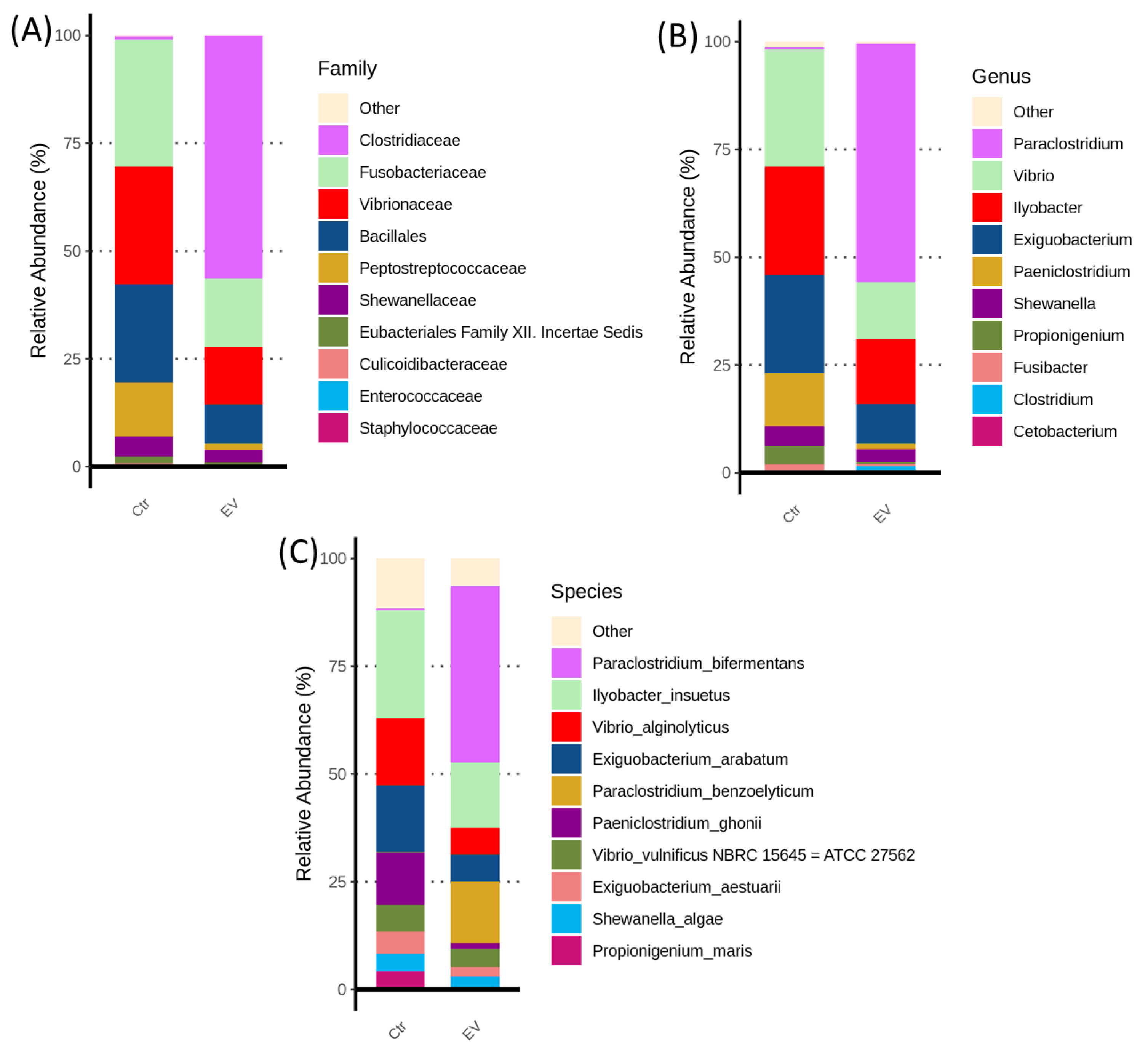
3.1. The Regulation of L. reuteri-Derived EVs on Microbiota in an In Vitro Cultured Intestinal Bacteria System
3.2. The Target Bacteria in an In Vitro Cultured Intestinal Bacteria System Using L. reuteri-Derived EVs Regulation
3.3. Prediction of Physiological and Growth-Related Genes of V. alginolyticus Affected by Small RNA Contained in L. reuteri-Derived EVs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fuller, R. Probiotics in man and animal. J. Appl. Bacteriol. 1989, 66, 365–378. [Google Scholar]
- Johnson-Henry, K.C.; Donato, K.A.; Shen-Tu, G.; Gordanpour, M.; Sherman, P.M. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-induced changes in epithelial barrier function. Infect. Immu. 2008, 76, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- Sha, H.; Li, L.; Lu, J.; Xiong, J. High nutrient induces virulence in the AHPND-causing Vibrio parahaemolyticus, interpretation from the ecological assembly of shrimp gut microbiota. Fish Shell. Immunol. 2022, 127, 758–765. [Google Scholar] [CrossRef]
- Rasheeda, M.K.; Rangamaran, V.R.; Srinivasan, S.; Ramaiah, S.K.; Gunasekaran, R.; Jaypal, S.; Gopal, D.; Ramalingam, K. Comparative profiling of microbial community of three economically important fishes reared in sea cages under tropical offshore environment. Mar. Genom. 2017, 34, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A.M. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Solmo salar L.). Aquaculture 2017, 467, 149–157. [Google Scholar] [CrossRef]
- Dawood, M.A.O.; Koshio, S.; Ishikawa, M.; Yokoyama, S.; El Basuini, M.F.; Hossain, M.S.; Nhu, T.H.; Dossou, S.; Moss, A.S. Effects of dietary supplementation of Lactobacillus rhamnosus or/and Lactococcus lactis on the growth, gut microbiota and immune responses of red sea bream, Pagrus major. Fish Shell. Immunol. 2016, 49, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhang, Z.; Wang, Y.; Li, S.; Ran, C.; Hu, J.; Xie, Y.; Li, W.; Zhou, Z. EPSP of L. casei BL23 protected against the infection caused by Aeromonas veronii via enhancement of immune response in zebrafish. Fr. Microbiol. 2017, 8, 2406. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Ran, C.; Qin, C.; Li, S.; Zhang, H.; de Vos, W.M.; Ringø, E.; Zhou, Z. Anti-infective effect of adhesive probiotic Lactobacillus in fish is correlated with their spatial distribution in the intestinal tissue. Sci. Rep. 2017, 7, 13195. [Google Scholar] [CrossRef]
- Lee, B.H.; Hu, Y.F.; Chu, Y.T.; Wu, Y.S.; Hsu, W.H.; Nan, F.H. Lactic acid bacteria-fermented diet containing bacterial extracellular vesicles inhibited pathogenic bacteria in Striped Beakfish (Oplegnathus fasciatus). Fermentation 2024, 10, 49. [Google Scholar] [CrossRef]
- Sha, Y.; Liu, M.; Wang, B.; Jiang, K.; Qi, C.; Wang, L. Bacterial population in intestines of Litopenaeus vannamei fed different probiotics or probiotic supernatant. J. Microbiol. Biotechnol. 2016, 26, 1736–1745. [Google Scholar]
- Lee, B.H.; Chen, Y.Z.; Shen, T.L.; Pan, T.M.; Hsu, W.H. Proteomic characterization of extracellular vesicles derived from lactic acid bacteria. Food Chem. 2023, 427, 136685. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.M.; Lee, B.H.; Hou, C.Y.; Hsu, W.H.; Tai, C.J. Probiotics-derived extracellular vesicles protect oxidative stress against H2O2 induction in placental cells. Fermentation 2022, 8, 74. [Google Scholar] [CrossRef]
- Lee, B.H.; Huang, S.C.; Hou, C.Y.; Chen, Y.Z.; Chen, Y.H.; Hazeena, S.H.; Hsu, W.H. Effect of polysaccharide derived from dehulled adlay on regulating gut microbiota and inhibiting Clostridioides difficile in an in vitro colonic fermentation model. Food Chem. 2023, 410, 135410. [Google Scholar] [CrossRef]
- Lee, B.H.; Wu, S.C.; Chien, H.Y.; Shen, T.L.; Hsu, W.H. Tomato-fruit-derived extracellular vesicles inhibit Fusobacterium nucleatum via lipid-mediated mechanism. Food Funct. 2023, 14, 8942. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.; Zhang, Z.; Cai, X.; Du, H.; Sun, G.; Cui, Y. An in vitro method to assess soil arsenic metabolism by human gut microbiota: Arsenic speciation and distribution. Environ. Sci. Technol. 2015, 49, 10675–10681. [Google Scholar] [CrossRef] [PubMed]
- Toyofuku, M.; Morinaga, K.; Hashimoto, Y.; Uhl, J.; Shimamura, H.; Inaba, H.; Schmitt-Kopplin, P.; Eberl, L.; Nomura, N. Membrane vesicle-mediated bacterial communication. ISME J. 2017, 11, 1504–1509. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, F.A.; Lecuyer, E. Small luggage for a long journey: Transfer of vesicle-enclosed small RNA in interspecies communication. Front. Microbiol. 2017, 8, 377. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Wu, S.C.; Shen, T.L.; Hsu, Y.Y.; Chen, C.H.; Hsu, W.H. The applications of Lactobacillus plantarum-derived extracellular vesicles as a novel natural antibacterial agent for improving quality and safety in tuna fish. Food Chem. 2021, 340, 128104. [Google Scholar] [CrossRef] [PubMed]
- Croatti, V.; Parolin, C.; Giordani, B.; Foschi, C.; Fedi, S.; Vitali, B. Lactobacilli extracellular vesicles: Potential postbiotics to support the vaginal microbiota homeostasis. Microb. Cell Fact. 2022, 21, 237. [Google Scholar] [CrossRef]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef]
- Zhou, L.; Han, F.; Lu, K.; Qiao, Y.; Li, E. Comparative study on prebiotic effects of different types of prebiotics in an in vitro fermentation by gut microbiota of shrimp (Litopenaeus vannamei). Aquaculture 2023, 574, 739687. [Google Scholar] [CrossRef]
- Wu, Y.S.; Chu, Y.T.; Chen, Y.Y.; Chang, C.S.; Lee, B.H.; Nan, F.H. Effects of dietary Lactobacillus reuteri and Pediococcus acidilactici on the cultured water qualities, the growth and non-specific immune responses of Penaeus vannamei. Fish Shell. Immunol. 2022, 127, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.T.; Hu, Y.F.; Lee, B.H.; Huang, C.Y.; Lin, Y.R.; Huang, S.N.; Chen, Y.Y.; Chang, J.J.; Nan, F.H. Dietary of Lactobacillus paracasei and Bifidobacterium longum improve non-specific immune responses, growth performance, and resistance against Vibrio parahaemolyticus in Penaeus vannamei. Fish Shell. Immunol. 2022, 128, 307–315. [Google Scholar] [CrossRef]
- Liu, C.H.; Wu, K.; Chu, T.W.; Wu, T.M. Dietary supplementation of probiotic, Bacillus subtilis E20, enhances the growth performance and disease resistance against Vibrio alginolyticus in parrot fish (Oplegnathus fasciatus). Aquac. Int. 2018, 26, 63–74. [Google Scholar] [CrossRef]
- Gastesoupe, F.J. Lactic acid bacteria increase the resistance of turbot larvae, Scophthalmus maximus, aginst pathogenic Vibrio. Aqu. Living Res. 1994, 7, 277–282. [Google Scholar] [CrossRef]
- Soh, M.; Tay, Y.C.; Lee, C.S.; Low, A.; Orban, L.; Jaafar, Z.; Seedorf, H. The intestinal digesta microbiota of tropical marine fish is largely uncultured and distinct from surrounding water microbiota. npj Biofilms Microb. 2024, 10, 11. [Google Scholar] [CrossRef]
- Suez, J.; Zmora, N.; Zilberman-Schapira, G.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Zur, M.; Regev-Lehavi, D.; Bez-Zeev Brik, R.; Federici, S.; et al. Reconsituion is impaired by probiotics and improved by autologous FMT. Cell 2018, 174, 1406–1423. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Ben-Zeev Brik, R.; et al. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell 2018, 174, 1388–1405. [Google Scholar] [CrossRef]
- MacDonald, I.A.; Kuehn, M.J. Offense and defense: Microbial membrane vesicles play both ways. Res. Microbiol. 2012, 163, 607–618. [Google Scholar] [CrossRef]
- Lee, J.H.; Choi, C.W.; Lee, T.; Kim, S.I.; Lee, J.C.; Shih, J.H. Transcription factor alpha plays an important role in the production of extracellular membrane-derived vesicles in Listeria monocytogenes. PLoS ONE 2013, 8, e73196. [Google Scholar]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Cañas, M.A.; Gimenez, R.; Fabrega, M.J.; Toloza, L.; Baldoma, L.; Badia, J. Outer membrane vesicles from the probiotic Escherichia coli Nissle1917 and the Commensal ECOR12 enter intestinal epithelial cells via clathrin dependent endocytosis and elicit differential effects on DNA Damage. PLoS ONE 2016, 11, e0160374. [Google Scholar] [CrossRef] [PubMed]
- Shenderov, B.A. Metabiotics: Novel idea or natural development of probiotic conception. Microb. Ecol. Health Dis. 2013, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Brotherton, D.; Inal, J. Communication is key: Extracellular vesicles as mediators of infection and defence during host-microbe interactions in animals and plant. FEMS Microbiol. Rev. 2022, 46, fuab044. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.H.; Chen, B.R.; Huang, C.T.; Lin, C.H. The immune activity of PT-peptide derived from anti-lipopolysaccharide factor of the swimming crab Portunus trituberculatus is enhanced when encapsulated in milk-derived extracellular vesicles. Mar. Drugs 2019, 17, 248. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Deng, Q.; Zhu, C.; Zhang, B. Application of extracellular vesicles in aquatic animals: A review of the lastest decade. Rev. Fish. Sci. Aquac. 2022, 30, 447–466. [Google Scholar] [CrossRef]
- Barathan, M.; Ng, S.L.; Lokanathan, Y.; Ng, M.H.; Law, J.X. Unseen weapons: Bacterial extracellular vesicles and the spread of antibiotic resistance in aquatic environments. Int. J. Mol. Sci. 2024, 25, 3080. [Google Scholar] [CrossRef]
- Bayat, F.; Afshar, A.; Baghban, N. Algal cells-derived extracellular vesicles: A review with special emphasis on their antimicrobial effects. Front. Microbiol. 2021, 12, 785716. [Google Scholar] [CrossRef] [PubMed]
- Schatz, D.; Schleyer, G.; Saltvedt, M.R.; Sandaa, R.A.; Feldmesser, E.; Vardi, A. Ecological significance of extracellular vesicles in modulating host-virus interactions during algal blooms. ISME J. 2021, 15, 3714–3721. [Google Scholar] [CrossRef]
- Chen, S.; Wang, H.; Katzianer, D.S.; Zhong, Z.; Zhu, J. LysR family activator-regulated major facilitator superfamily transporters are involved in Vibrio cholerae antimicrobial compound resistance and intestinal colonization. Int. J. Antimicrob. Agents 2013, 41, 188–192. [Google Scholar] [CrossRef]
- Kovacikova, G.; Lin, W.; Skorupski, K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J. Bacteriol. 2010, 192, 4181–4191. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Bartlam, M.; Zeng, Q.; Miyatake, H.; Hisano, T.; Miki, K.; Wong, L.L.; Gao, G.F.; Rao, Z. Crystal structure of human pirin: An iron-binding nuclear protein and transcription cofactor. J. Biol. Chem. 2004, 279, 1491–1498. [Google Scholar] [CrossRef] [PubMed]
- Soo, P.C.; Horng, Y.T.; Lai, M.J.; Wei, J.R.; Hsieh, S.C.; Chang, Y.L.; Tsai, Y.H.; Lai, H.C. Pirin regulates pyruvate catabolism by interacting with pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J. Bacteriol. 2007, 189, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ghai, I.; Bajaj, H.; Arun Bafna, J.; El Damrany Hussein, H.A.; Winterhalter, M.; Wagner, R. Ampicillin permeation across OmpF, the major outer-membrane channel in Escherichia coli. J. Biol. Chem. 2018, 293, 7030–7037. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Huang, D.; Zhou, Q.; Li, F.; Tan, X.; Wang, J.; Wang, X. The influence of outer membrane protein on ampicillin resistance of Vibrio parahaemolyticus. Can. J. Infect. Dis. Med. Microbiol. 2023, 2023, 8079091. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Huang, Y.; Liu, P.; Yan, L.; Zhou, Y.; Yang, C.; Wu, Y.; Qin, J.; Guo, Y.; Pei, X.; et al. Population genomics of the food-borne pathogen Vibrio fluvialis reveals lineage associated pathogenicity-related genetic elements. Microb. Genom. 2022, 8, 00076. [Google Scholar] [CrossRef] [PubMed]
- Restrepo-Benavides, M.; Lozano-Arce, D.; Gonzalez-Garcia, L.N.; Báez-Aguirre, F.; Ariza-Aranguren, G.; Faccini, D.; Zambrano, M.M.; Jiménez, P.; Fernández-Bravo, A.; Restrepo, S.; et al. Unveiling potential virulence determinants in Vibrio isolates from Anadara tuberculosa through whole genome analyses. Microbiol. Spectr. 2024, 12, e02928-23. [Google Scholar] [CrossRef] [PubMed]
- Blanco, J.; Moore, R.A.; Aran, V.K.; Viola, R.E. A structural basis for the mechanism of asparate-β-semialdehyde dehydrogenase from Vibrio cholerae. Protein Sci. 2013, 12, 27–33. [Google Scholar] [CrossRef]
- Marrero, K.; Marrero, E.; Silva, Y.; Rodriguez, B.; Campos, J.; Ledon, T.; Valle, E.; Fando, R. The genome of Vibrio cholerae contains two different and functional genes for aspartate semialdehyde dehydrogenases. Rev. CENIC Cienc. Biológicas 2004, 35, 163–176. [Google Scholar]





| Gene ID | Gene Name | Small RNA (Reads) |
|---|---|---|
| NAL94_RS00020 | LysR | 255 |
| NAL94_RS04885 | pirin | 1700 |
| NAL94_RS06120 | MIpA/OmpV | 348 |
| NAL94_RS07195 | CatB | 571 |
| NAL94_RS12345 | aspartate-semlaldehyde dehydrogenase | 2020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, B.-H.; Hu, Y.-F.; Das, S.P.; Chu, Y.-T.; Hsu, W.-H.; Nan, F.-H. An In Vitro System Mimics the Intestinal Microbiota of Striped Beakfish (Oplegnathus fasciatus) and Inhibits Vibrio alginolyticus by Limosilactobacillus reuteri-Derived Extracellular Vesicles. Animals 2024, 14, 1792. https://doi.org/10.3390/ani14121792
Lee B-H, Hu Y-F, Das SP, Chu Y-T, Hsu W-H, Nan F-H. An In Vitro System Mimics the Intestinal Microbiota of Striped Beakfish (Oplegnathus fasciatus) and Inhibits Vibrio alginolyticus by Limosilactobacillus reuteri-Derived Extracellular Vesicles. Animals. 2024; 14(12):1792. https://doi.org/10.3390/ani14121792
Chicago/Turabian StyleLee, Bao-Hong, Yeh-Fang Hu, Sofia Priyadarsani Das, Yu-Ting Chu, Wei-Hsuan Hsu, and Fan-Hua Nan. 2024. "An In Vitro System Mimics the Intestinal Microbiota of Striped Beakfish (Oplegnathus fasciatus) and Inhibits Vibrio alginolyticus by Limosilactobacillus reuteri-Derived Extracellular Vesicles" Animals 14, no. 12: 1792. https://doi.org/10.3390/ani14121792






