Cryptosporidium spp. Infection and Genotype Identification in Pre-Weaned and Post-Weaned Calves in Yunnan Province, China
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection
2.2. DNA Extraction and PCR Amplification
2.3. Sequence Analysis and Phylogenetic Tree
2.4. Statistical Analysis
3. Results
3.1. Prevalence of Cryptosporidium spp.
3.2. Genotyping of Cryptosporidium spp.
3.3. Subtyping of Cryptosporidium spp.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chalmers, R.M.; Davies, A.P.; Tyler, K. Cryptosporidium. Microbiology 2019, 165, 500–502. [Google Scholar] [CrossRef]
- Yang, X.; Guo, Y.; Xiao, L.; Feng, Y. Molecular Epidemiology of Human Cryptosporidiosis in Low- and Middle-Income Countries. Clin. Microbiol. Rev. 2021, 34, e00087-19. [Google Scholar] [CrossRef]
- Liu, A.; Gong, B.; Liu, X.; Shen, Y.; Wu, Y.; Zhang, W.; Cao, J. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018). PLoS Neglected Trop. Dis. 2020, 14, e0008146. [Google Scholar] [CrossRef]
- Lv, S.; Tian, L.G.; Liu, Q.; Qian, M.B.; Fu, Q.; Steinmann, P.; Chen, J.X.; Yang, G.J.; Yang, K.; Zhou, X.N. Water-related parasitic diseases in China. Int. J. Environ. Res. Public Health 2013, 10, 1977–2016. [Google Scholar] [CrossRef]
- Thomson, S.; Hamilton, C.A.; Hope, J.C.; Katzer, F.; Mabbott, N.A.; Morrison, L.J.; Innes, E.A. Bovine cryptosporidiosis: Impact, host-parasite interaction and control strategies. Vet. Res. 2017, 48, 42. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef]
- Santín, M. Clinical and subclinical infections with Cryptosporidium in animals. N. Z. Vet. J. 2013, 61, 1–10. [Google Scholar] [CrossRef]
- Shaw, H.J.; Innes, E.A.; Morrison, L.J.; Katzer, F.; Wells, B. Long-term production effects of clinical cryptosporidiosis in neonatal calves. Int. J. Parasitol. 2020, 50, 371–376. [Google Scholar] [CrossRef]
- Jacobson, C.; Al-Habsi, K.; Ryan, U.; Williams, A.; Anderson, F.; Yang, R.; Abraham, S.; Miller, D. Cryptosporidium infection is associated with reduced growth and diarrhoea in goats beyond weaning. Vet. Parasitol. 2018, 260, 30–37. [Google Scholar] [CrossRef]
- Santin, M. Cryptosporidium and Giardia in Ruminants. Vet. Clin. N. Am. Food. Anim. Pract. 2020, 36, 223–238. [Google Scholar] [CrossRef]
- Li, N.; Zhao, W.; Song, S.; Ye, H.; Chu, W.; Guo, Y.; Feng, Y.; Xiao, L. Diarrhoea outbreak caused by coinfections of Cryptosporidium parvum subtype IIdA20G1 and rotavirus in pre-weaned dairy calves. Transbound Emerg. Dis. 2022, 69, e1606–e1607. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, R.; Cai, M.; Jiang, W.; Feng, Y.; Xiao, L. Outbreak of cryptosporidiosis due to Cryptosporidium parvum subtype IIdA19G1 in neonatal calves on a dairy farm in China. Int. J. Parasitol. 2019, 49, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Wang, R.; Huang, J.; Wang, H.; Zhao, J.; Luo, N.; Li, J.; Zhang, Z.; Zhang, L. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in Northwestern China. Parasit. Vectors 2014, 7, 529. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Su, D.; Meng, X.; Liang, R.; Wang, W.; Li, N.; Guo, Y.; Guo, A.; Li, S.; Zhao, Z.; et al. Cryptosporidiosis outbreak caused by Cryptosporidium parvum subtype IIdA20G1 in neonatal calves. Transbound Emerg. Dis. 2022, 69, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Feng, Y.; Xiao, L. An Update on Zoonotic Cryptosporidium Species and Genotypes in Humans. Animals 2021, 11, 3307. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Cao, X.F.; Deng, L.; Li, W.; Huang, X.M.; Lan, J.C.; Xiao, Q.C.; Zhong, Z.J.; Feng, F.; Zhang, Y.; et al. Epidemiology of Cryptosporidium infection in cattle in China: A review. Parasite 2017, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Huang, J.; Qin, H.; Wang, L.; Li, J.; Zhang, L. Cryptosporidium parvum and gp60 genotype prevalence in dairy calves worldwide: A systematic review and meta-analysis. Acta Trop. 2023, 240, 106843. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Xiao, L. Molecular Epidemiology of Cryptosporidiosis in China. Front. Microbiol. 2017, 8, 1701. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gong, X.; Zhu, K.; Li, N.; Yu, Z.; Guo, Y.; Weng, Y.; Kvac, M.; Feng, Y.; Xiao, L. Prevalence and genotypic identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in pre-weaned dairy calves in Guangdong, China. Parasit. Vectors 2019, 12, 41. [Google Scholar] [CrossRef]
- Wang, R.; Ma, G.; Zhao, J.; Lu, Q.; Wang, H.; Zhang, L.; Jian, F.; Ning, C.; Xiao, L. Cryptosporidium andersoni is the predominant species in post-weaned and adult dairy cattle in China. Parasitol. Int. 2011, 60, 1–4. [Google Scholar] [CrossRef]
- Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010, 124, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Ortega, Y.; He, G.; Das, P.; Xu, M.; Zhang, X.; Fayer, R.; Gatei, W.; Cama, V.; Xiao, L. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 2007, 144, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Shaik, J.S.; Grigg, M.E. Genomics and molecular epidemiology of Cryptosporidium species. Acta Trop. 2018, 184, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Strong, W.B.; Gut, J.; Nelson, R.G. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 2000, 68, 4117–4134. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017, 8–9, 14–32. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wan, M.; Yang, F.; Li, N.; Xiao, L.; Feng, Y.; Guo, Y. Development and Application of a gp60-Based Subtyping Tool for Cryptosporidium bovis. Microorganisms 2021, 9, 2067. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Huang, N.; Jiang, W.; Wang, X.; Li, N.; Guo, Y.; Kvac, M.; Feng, Y.; Xiao, L. Subtyping Cryptosporidium ryanae: A Common Pathogen in Bovine Animals. Microorganisms 2020, 8, 1107. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yang, W.; Ryan, U.; Zhang, L.; Kvac, M.; Koudela, B.; Modry, D.; Li, N.; Fayer, R.; Xiao, L. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J. Clin. Microbiol. 2011, 49, 34–41. [Google Scholar] [CrossRef]
- Cho, Y.I.; Yoon, K.J. An overview of calf diarrhea-infectious etiology, diagnosis, and intervention. J. Vet. Sci. 2014, 15, 1–17. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, T.; Koehler, A.V.; Hu, M.; Gasser, R.B. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei Province, China. Parasit. Vectors 2017, 10, 519. [Google Scholar] [CrossRef]
- Gao, H.; Liang, G.; Su, N.; Li, Q.; Wang, D.; Wang, J.; Zhao, L.; Kang, X.; Guo, K. Prevalence and Molecular Characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Diarrheic and Non-Diarrheic Calves from Ningxia, Northwestern China. Animals 2023, 13, 1983. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Liu, Z.; Yan, F.; Zhang, Z.; Zhang, G.; Zhang, L.; Jian, F.; Zhang, S.; Ning, C.; Wang, R. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet. Parasitol. 2017, 248, 68–73. [Google Scholar] [CrossRef]
- Hu, S.; Wan, M.; Huang, W.; Wang, W.; Liang, R.; Su, D.; Li, N.; Xiao, L.; Feng, Y.; Guo, Y. Age and episode-associated occurrence of Cryptosporidium species and subtypes in a birth-cohort of dairy calves. Transbound Emerg. Dis. 2022, 69, e1710–e1720. [Google Scholar] [CrossRef]
- Li, S.; Zou, Y.; Wang, P.; Qu, M.R.; Zheng, W.B.; Wang, P.; Chen, X.Q.; Zhu, X.Q. Prevalence and multilocus genotyping of Cryptosporidium spp. in cattle in Jiangxi Province, southeastern China. Parasitol. Res. 2021, 120, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.X.; Zou, Y.; Li, T.S.; Chen, H.; Wang, S.S.; Cao, F.Q.; Yang, J.F.; Sun, X.L.; Zhu, X.Q.; Zou, F.C. First report of the prevalence and genetic characterization of Giardia duodenalis and Cryptosporidium spp. in Yunling cattle in Yunnan Province, southwestern China. Microb. Pathog. 2021, 158, 105025. [Google Scholar] [CrossRef]
- Qin, H.; Lang, J.; Zhang, K.; Zhang, A.; Chen, Y.; Fu, Y.; Wang, C.; Zhang, L. Study on genetic characteristics of Cryptosporidium isolates and first report of C. parvum IIdA24G2 subtype in dairy cattle in China. Parasitol. Res. 2024, 123, 81. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.; Chang, Y.; Yu, F.; Zhang, S.; Wang, R.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 2020, 27, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Dan, J.; Yan, G.; Tu, R.; Tian, Y.; Cao, S.; Shen, L.; Deng, J.; Yu, S.; Geng, Y.; et al. Occurrence and genotyping of Giardia duodenalis and Cryptosporidium in pre-weaned dairy calves in central Sichuan province, China. Parasite 2018, 25, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Wu, Y.; Jing, B.; Xu, C.; Chen, Y.; Yu, F.; Wei, Z.; Zhang, Y.; Cui, Z.; Qi, M.; et al. Seasonal monitoring of Cryptosporidium species and their genetic diversity in neonatal calves on two large-scale farms in Xinjiang, China. J. Eukaryot Microbiol. 2022, 69, e12878. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, G.; Gong, Y.; Zhang, L. Advances and Perspectives on the Epidemiology of Bovine Cryptosporidium in China in the Past 30 Years. Front. Microbiol. 2017, 8, 1823. [Google Scholar] [CrossRef]
- Meng, Y.W.; Shu, F.F.; Pu, L.H.; Zou, Y.; Yang, J.F.; Zou, F.C.; Zhu, X.Q.; Li, Z.; He, J.J. Occurrence and Molecular Characterization of Cryptosporidium spp. in Dairy Cattle and Dairy Buffalo in Yunnan Province, Southwest China. Animals 2022, 12, 1031. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Escalante, L.; Yang, C.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999, 65, 1578–1583. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Li, N.; Duan, L.; Xiao, L. Cryptosporidium genotype and subtype distribution in raw wastewater in Shanghai, China: Evidence for possible unique Cryptosporidium hominis transmission. J. Clin. Microbiol. 2009, 47, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Hatam-Nahavandi, K.; Ahmadpour, E.; Carmena, D.; Spotin, A.; Bangoura, B.; Xiao, L. Cryptosporidium infections in terrestrial ungulates with focus on livestock: A systematic review and meta-analysis. Parasit. Vectors 2019, 12, 453. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zhang, N.Z.; Gong, Q.L.; Zhao, Q.; Zhang, X.X. Prevalence of Cryptosporidium in dairy cattle in China during 2008-2018: A systematic review and meta-analysis. Microb. Pathog. 2019, 132, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, H.; Sun, Y.; Zhang, L.; Jian, F.; Qi, M.; Ning, C.; Xiao, L. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan, China. J. Clin. Microbiol. 2011, 49, 1077–1082. [Google Scholar] [CrossRef]
- Cai, M.; Guo, Y.; Pan, B.; Li, N.; Wang, X.; Tang, C.; Feng, Y.; Xiao, L. Longitudinal monitoring of Cryptosporidium species in pre-weaned dairy calves on five farms in Shanghai, China. Vet. Parasitol. 2017, 241, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, P.; Zhao, X.; Xu, H.; Wu, W.; Wang, Y.; Guo, Y.; Wang, L.; Feng, Y.; Xiao, L. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015, 207, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.Z.; Fang, Y.Q.; Wang, X.T.; Zhang, L.X.; Wang, R.J.; Du, S.Z.; Guo, Y.X.; Jia, Y.Q.; Yao, L.; Liu, Q.D.; et al. Molecular characterization of Cryptosporidium spp. in pre-weaned calves in Shaanxi Province, north-western China. J. Med. Microbiol. 2015, 64, 111–116. [Google Scholar] [CrossRef]
- Zhang, X.X.; Tan, Q.D.; Zhou, D.H.; Ni, X.T.; Liu, G.X.; Yang, Y.C.; Zhu, X.Q. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol. Res. 2015, 114, 2781–2787. [Google Scholar] [CrossRef]
- Huang, J.; Yue, D.; Qi, M.; Wang, R.; Zhao, J.; Li, J.; Shi, K.; Wang, M.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet. Res. 2014, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Wang, H.; Jing, B.; Wang, D.; Wang, R.; Zhang, L. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet. Parasitol. 2015, 212, 404–407. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, H.; Zhang, Z.; Li, J.; Wang, C.; Zhao, J.; Hu, S.; Wang, R.; Zhang, L.; Wang, M. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Beijing, China. Vet. Parasitol. 2016, 219, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Wu, Y.; Sun, M.; Chang, Y.; Lin, X.; Yu, L.; Hu, S.; Zhang, X.; Zheng, S.; Cui, Z.; et al. Molecular epidemiology of Cryptosporidium spp. in dairy cattle in Guangdong Province, South China. Parasitology 2019, 146, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Chai, H.L.; Wang, M.Y.; Zhang, Z.S.; Han, W.X.; Yang, B.; Wang, Y.; Zhang, S.; Zhao, W.H.; Ma, Y.M.; et al. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle in Central Inner Mongolia, Northern China. Vet. Res. 2023, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, D.A.; Ola-Fadunsin, S.D.; Ruviniyia, K.; Gimba, F.I.; Chandrawathani, P.; Lim, Y.A.L.; Jesse, F.F.A.; Sharma, R.S.K. Molecular detection and epidemiological risk factors associated with Cryptosporidium infection among cattle in Peninsular Malaysia. Food Waterborne Parasitol. 2019, 14, e00035. [Google Scholar] [CrossRef]
- Guy, R.A.; Yanta, C.A.; Bauman, C.A. Molecular identification of Cryptosporidium species in Canadian post-weaned calves and adult dairy cattle. Vet. Parasitol. Reg. Stud. Rep. 2022, 34, 100777. [Google Scholar] [CrossRef] [PubMed]
- Jang, D.H.; Cho, H.C.; Shin, S.U.; Kim, E.M.; Park, Y.J.; Hwang, S.; Park, J.; Choi, K.S. Prevalence and distribution pattern of Cryptosporidium spp. among pre-weaned diarrheic calves in the Republic of Korea. PLoS ONE 2021, 16, e0259824. [Google Scholar] [CrossRef] [PubMed]
- Keomoungkhoun, B.; Arjentinia, I.; Sangmaneedet, S.; Taweenan, W. Molecular prevalence and associated risk factors of Cryptosporidium spp. infection in dairy cattle in Khon Kaen, Thailand. Vet. World 2024, 17, 371–378. [Google Scholar] [CrossRef]
- Holzhausen, I.; Lendner, M.; Gohring, F.; Steinhofel, I.; Daugschies, A. Distribution of Cryptosporidium parvum gp60 subtypes in calf herds of Saxony, Germany. Parasitol. Res. 2019, 118, 1549–1558. [Google Scholar] [CrossRef]
- Benito, A.A.; Monteagudo, L.V.; Arnal, J.L.; Baselga, C.; Quílez, J. Occurrence and genetic diversity of rotavirus A in faeces of diarrheic calves submitted to a veterinary laboratory in Spain. Prev. Vet. Med. 2020, 185, 105196. [Google Scholar] [CrossRef]
- Oliveira, J.S.; Martins, F.D.C.; Ladeia, W.A.; Cortela, I.B.; Valadares, M.F.; Matos, A.; Caldart, E.T.; Ayres, H.; Navarro, I.T.; Freire, R.L. Identification, molecular characterization and factors associated with occurrences of Cryptosporidium spp. in calves on dairy farms in Brazil. Rev. Bras. Parasitol. Vet. 2021, 30, e009621. [Google Scholar] [CrossRef]
- Díaz, P.; Varcasia, A.; Pipia, A.P.; Tamponi, C.; Sanna, G.; Prieto, A.; Ruiu, A.; Spissu, P.; Díez-Baños, P.; Morrondo, P.; et al. Molecular characterisation and risk factor analysis of Cryptosporidium spp. in calves from Italy. Parasitol. Res. 2018, 117, 3081–3090. [Google Scholar] [CrossRef] [PubMed]
- Elmahallawy, E.K.; Sadek, H.A.; Aboelsoued, D.; Aloraini, M.A.; Alkhaldi, A.A.M.; Abdel-Rahman, S.M.; Bakir, H.Y.; Arafa, M.I.; Hassan, E.A.; Elbaz, E.; et al. Parasitological, Molecular, and Epidemiological Investigation of Cryptosporidium Infection Among Cattle and Buffalo Calves From Assiut Governorate, Upper Egypt: Current Status and Zoonotic Implications. Front. Vet. Sci. 2022, 9, 899854. [Google Scholar] [CrossRef] [PubMed]
- Gattan, H.S.; Alshammari, A.; Marzok, M.; Salem, M.; Al-Jabr, O.A.; Selim, A. Prevalence of Cryptosporidium infection and associated risk factors in calves in Egypt. Sci. Rep. 2023, 13, 17755. [Google Scholar] [CrossRef]
- El-Ashram, S.; Aboelhadid, S.M.; Kamel, A.A.; Mahrous, L.N.; Abdelwahab, K.H. Diversity of Parasitic Diarrhea Associated with Buxtonella Sulcata in Cattle and Buffalo Calves with Control of Buxtonellosis. Animals 2019, 9, 259. [Google Scholar] [CrossRef]
- Maurya, P.S.; Rakesh, R.L.; Pradeep, B.; Kumar, S.; Kundu, K.; Garg, R.; Ram, H.; Kumar, A.; Banerjee, P.S. Prevalence and risk factors associated with Cryptosporidium spp. infection in young domestic livestock in India. Trop. Animal Health Prod. 2013, 45, 941–946. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.; Juyal, P.; Singla, L. Prevalence of cryptosporidiosis in neonatal buffalo calves in Ludhiana district of Punjab, India. Asian J. Animal Vet. Adv. 2012, 7, 512–520. [Google Scholar] [CrossRef]
- Geng, H.L.; Ni, H.B.; Li, J.H.; Jiang, J.; Wang, W.; Wei, X.Y.; Zhang, Y.; Sun, H.T. Prevalence of Cryptosporidium spp. in Yaks (Bos grunniens) in China: A Systematic Review and Meta-Analysis. Front. Cell Infect. Microbiol. 2021, 11, 770612. [Google Scholar] [CrossRef]
- Guo, Y.; Ryan, U.; Feng, Y.; Xiao, L. Association of Common Zoonotic Pathogens with Concentrated Animal Feeding Operations. Front. Microbiol. 2021, 12, 810142. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, K.; Zhang, Y.; Jing, B.; Chen, Y.; Xu, C.; Wang, T.; Qi, M.; Zhang, L. Genetic Diversity of Cryptosporidium parvum in Neonatal Dairy Calves in Xinjiang, China. Pathogens 2020, 9, 692. [Google Scholar] [CrossRef] [PubMed]
- Feltus, D.C.; Giddings, C.W.; Khaitsa, M.L.; McEvoy, J.M. High prevalence of Cryptosporidium bovis and the deer-like genotype in calves compared to mature cows in beef cow-calf operations. Vet. Parasitol. 2008, 151, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Silverlås, C.; de Verdier, K.; Emanuelson, U.; Mattsson, J.G.; Björkman, C. Cryptosporidium infection in herds with and without calf diarrhoeal problems. Parasitol. Res. 2010, 107, 1435–1444. [Google Scholar] [CrossRef] [PubMed]
- Murakoshi, F.; Xiao, L.; Matsubara, R.; Sato, R.; Kato, Y.; Sasaki, T.; Fukuda, Y.; Tada, C.; Nakai, Y. Molecular characterization of Cryptosporidium spp. in grazing beef cattle in Japan. Vet. Parasitol. 2012, 187, 123–128. [Google Scholar] [CrossRef]
- Abeywardena, H.; Jex, A.R.; Koehler, A.V.; Rajapakse, R.P.; Udayawarna, K.; Haydon, S.R.; Stevens, M.A.; Gasser, R.B. First molecular characterization of Cryptosporidium and Giardia from bovines (Bos taurus and Bubalus bubalis) in Sri Lanka: Unexpected absence of C. parvum from pre-weaned calves. Parasit. Vectors 2014, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.T.; Fukuda, Y.; Tada, C.; Sato, R.; Duong, B.; Nguyen, D.T.; Nakai, Y. Molecular characterization of Cryptosporidium in native beef calves in central Vietnam. Parasitol. Res. 2012, 111, 1817–1820. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ryan, U.; Feng, Y.; Xiao, L. Emergence of zoonotic Cryptosporidium parvum in China. Trends Parasitol. 2022, 38, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.H.; Ren, W.X.; Gao, M.; Bian, Q.Q.; Hu, B.; Cong, M.M.; Lin, Q.; Wang, R.J.; Qi, M.; Qi, M.Z.; et al. Genotyping Cryptosporidium andersoni in cattle in Shaanxi Province, Northwestern China. PLoS ONE 2013, 8, e60112. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, H.; Zhao, X.; Zhang, L.; Zhang, G.; Guo, M.; Liu, L.; Feng, Y.; Xiao, L. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J. Clin. Microbiol. 2013, 51, 557–563. [Google Scholar] [CrossRef]
- Misic, Z.; Abe, N. Subtype analysis of Cryptosporidium parvum isolates from calves on farms around Belgrade, Serbia and Montenegro, using the 60 kDa glycoprotein gene sequences. Parasitology 2007, 134, 351–358. [Google Scholar] [CrossRef]
- Kabir, M.H.B.; Ceylan, O.; Ceylan, C.; Shehata, A.A.; Bando, H.; Essa, M.I.; Xuan, X.; Sevinc, F.; Kato, K. Molecular detection of genotypes and subtypes of Cryptosporidium infection in diarrheic calves, lambs, and goat kids from Turkey. Parasitol. Int. 2020, 79, 102163. [Google Scholar] [CrossRef]
- Taha, S.; Elmalik, K.; Bangoura, B.; Lendner, M.; Mossaad, E.; Daugschies, A. Molecular characterization of bovine Cryptosporidium isolated from diarrheic calves in the Sudan. Parasitol. Res. 2017, 116, 2971–2979. [Google Scholar] [CrossRef] [PubMed]
- Qi, M.; Cai, J.; Wang, R.; Li, J.; Jian, F.; Huang, J.; Zhou, H.; Zhang, L. Molecular characterization of Cryptosporidium spp. and Giardia duodenalis from yaks in the central western region of China. Microbiology 2015, 15, 108. [Google Scholar] [CrossRef] [PubMed]
- Boughattas, S.; Behnke, J.M.; Al-Ansari, K.; Sharma, A.; Abu-Alainin, W.; Al-Thani, A.; Abu-Madi, M.A. Molecular Analysis of the Enteric Protozoa Associated with Acute Diarrhea in Hospitalized Children. Front. Cell. Infect. Microbiol. 2017, 7, 343. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.M.; Smith, R.P.; Hadfield, S.J.; Elwin, K.; Giles, M. Zoonotic linkage and variation in Cryptosporidium parvum from patients in the United Kingdom. Parasitol. Res. 2011, 108, 1321–1325. [Google Scholar] [CrossRef] [PubMed]
- de Lucio, A.; Merino, F.J.; Martínez-Ruiz, R.; Bailo, B.; Aguilera, M.; Fuentes, I.; Carmena, D. Molecular genotyping and sub-genotyping of Cryptosporidium spp. isolates from symptomatic individuals attending two major public hospitals in Madrid, Spain. Infect. Genet. Evol. 2016, 37, 49–56. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Hira, P.R.; Zhou, L.; Al-Ali, F.M.; Al-Shelahi, F.A.; Shweiki, H.M.; Iqbal, J.; Khalid, N.; Xiao, L. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 2005, 43, 2805–2809. [Google Scholar] [CrossRef]
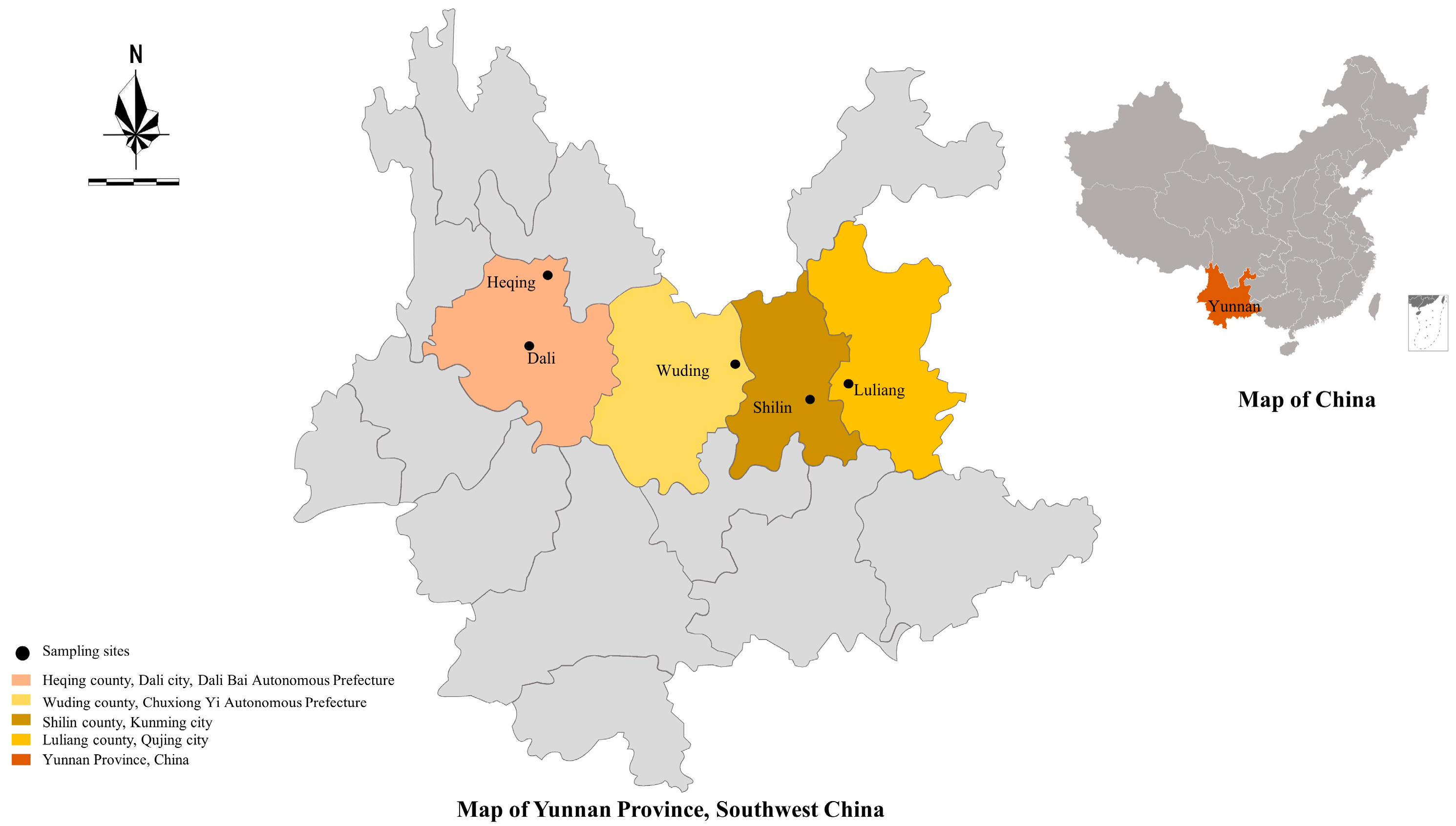
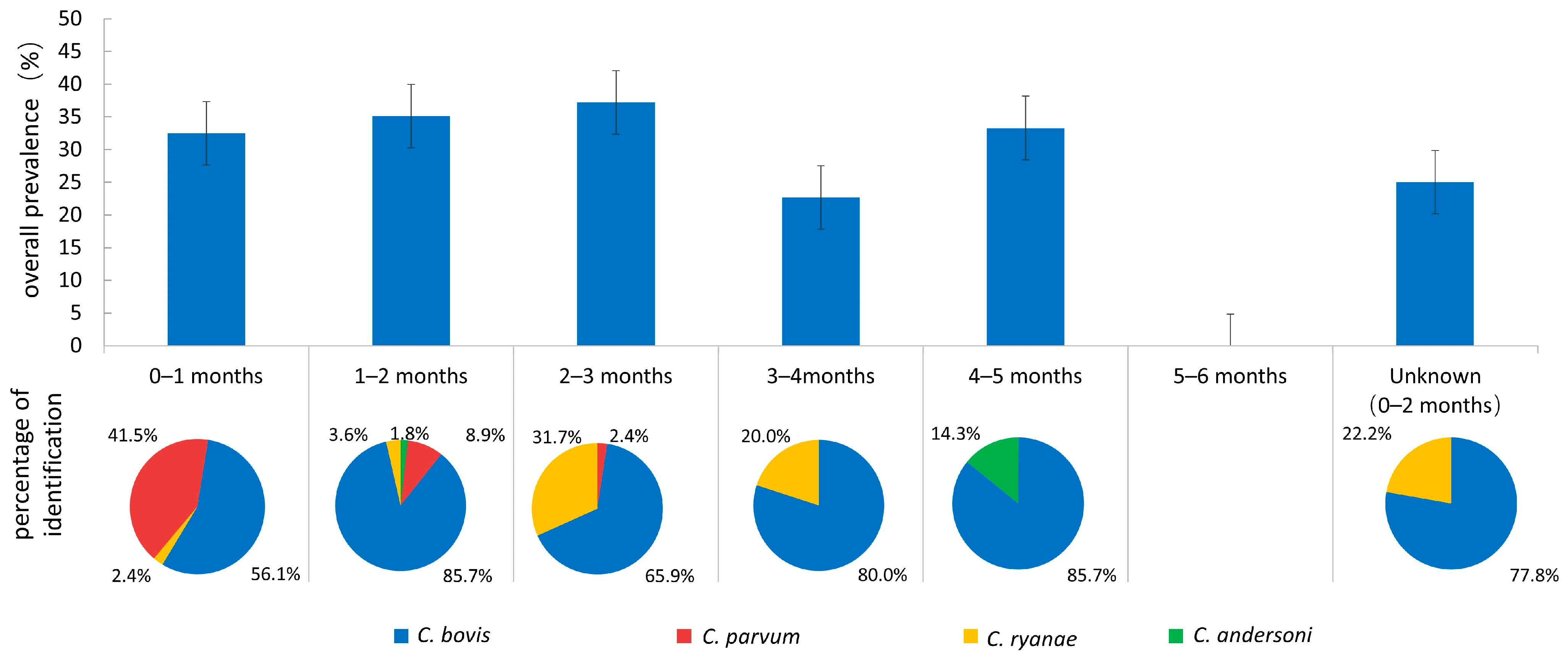
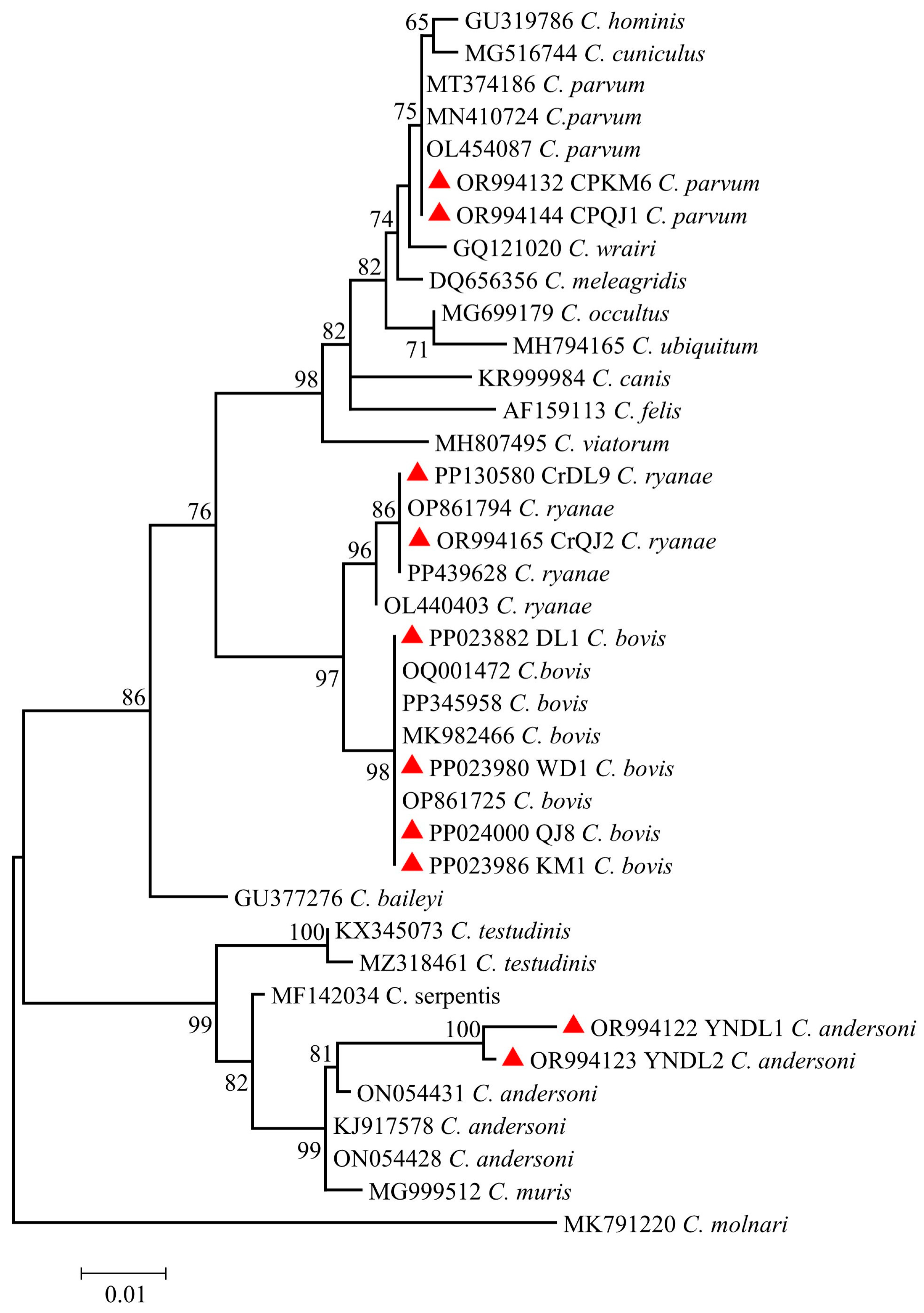
| Variable | Category | Sample Size | No. Positive | Prevalence (%) (95%CI) | OR (95%CI) | p-Value |
|---|---|---|---|---|---|---|
| Region | Kunming | 53 | 24 | 45.3 (31.43–59.13) | 5.68 (2.60–12.37) | <0.01 |
| Dali | 312 | 120 | 38.5 (33.00–43.90) | 4.29 (2.34–7.85) | ||
| Chuxiong | 23 | 6 | 26.1 (6.67–45.50) | 2.42 (0.82–7.17) | ||
| Qujing | 110 | 14 | 12.7 (6.40–19.05) | Reference | ||
| Sex | Female | 392 | 127 | 32.4 (27.74–37.05) | Reference | 0.63 |
| Male | 106 | 37 | 34.9 (25.68–44.13) | 1.12 (0.71–1.76) | ||
| Age | Pre-weaned (0–60 days) | 316 | 106 | 33.5 (28.31–38.78) | 1.08 (0.73–1.59) | 0.70 |
| Post-weaned (61–180 days) | 182 | 58 | 31.9 (25.03–38.70) | Reference | ||
| Total | 498 | 164 | 32.9 (28.79–37.07) | |||
| Factor | Category | Cryptosporidium Species | Cryptosporidium Subtypes | ||
|---|---|---|---|---|---|
| C. bovis | C. parvum | C. ryanae | |||
| Region | Dali | C. bovis (98), C. ryanae (17), C. parvum (3), C. andersoni (2) | XXVIb (22), XXVIe (17), XXVIf (7), XXVId (3), XXVIc (2) , XXVIa (1) | – | XXIf (2), XXId (1), XXIe (1), XXIg (1) |
| Kunming | C. parvum (17), C. bovis (7) | XXVIb (4), XXVIa (1) | IIdA19G1(7), IIdA18G1 (3) | – | |
| Qujing | C. bovis (8), C. parvum (3), C. ryanae (3) | XXVIa (1) | IIdA18G1 (2) | – | |
| Chuxiong | C. bovis (6) | XXVIb (18),XXVIc (1), XXVIe (1) | – | ||
| Sex | Female | C. bovis (84), C. parvum (23), C. ryanae (18), C. andersoni (2) | XXVIe (15), XXVIa (3), XXVIc (3), XXVId (2), XXVIf (1) | IIdA19G1 (7), IIdA18G1 (5) | XXIf (2), XXIe (1), XXIg (1) |
| Male | C. bovis (35), C. ryanae (2) | XXVIb(8), XXVIf (6), XXVIe (3), XXVId (1) | – | XXId (1) | |
| Age | Pre-weaned (0–60 days) | C. bovis (78), C. parvum (22), C. ryanae (5), C. andersoni (1) | XXVIb (20), XXVIe (14), XXVIf (4), XXVIa (3), XXVIc (1), XXVId (1) | IIdA19G1 (7), IIdA18G1 (5) | XXId (1), XXIf (1) |
| Post-weaned (61–180 days) | C. bovis (41), C. ryanae (15), C. andersoni (1), C. parvum (1) | XXVIb (6), XXVIe (4), XXVIf (3), XXVIc (2), XXVId (2) | – | XXIe (1), XXIf (1), XXIg (1) | |
| Total | C. bovis (119), C. parvum (23), C. ryanae (20), C. andersoni (2) | XXVIb (26), XXVIe (18), XXVIf (7), XXVIa (3), XXVIc (3), XXVId (3) | IIdA19G1 (7), IIdA18G1 (5) | XXIf(2), XXId (1), XXIe (1), XXIg (1) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, M.-L.; Heng, Z.-J.; Li, L.-J.; Yang, J.-F.; He, J.-J.; Zou, F.-C.; Shu, F.-F. Cryptosporidium spp. Infection and Genotype Identification in Pre-Weaned and Post-Weaned Calves in Yunnan Province, China. Animals 2024, 14, 1907. https://doi.org/10.3390/ani14131907
Deng M-L, Heng Z-J, Li L-J, Yang J-F, He J-J, Zou F-C, Shu F-F. Cryptosporidium spp. Infection and Genotype Identification in Pre-Weaned and Post-Weaned Calves in Yunnan Province, China. Animals. 2024; 14(13):1907. https://doi.org/10.3390/ani14131907
Chicago/Turabian StyleDeng, Meng-Ling, Zhao-Jun Heng, Liu-Jia Li, Jian-Fa Yang, Jun-Jun He, Feng-Cai Zou, and Fan-Fan Shu. 2024. "Cryptosporidium spp. Infection and Genotype Identification in Pre-Weaned and Post-Weaned Calves in Yunnan Province, China" Animals 14, no. 13: 1907. https://doi.org/10.3390/ani14131907





