Genome-Wide Association Study for Weight Loss at the End of Dry-Curing of Hams Produced from Purebred Heavy Pigs
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Trial
2.2. Measurement of Ham Weight Loss at the End of Dry-Curing
2.3. Genotyping
2.4. Genetic Parameters and Genome-Wide Association Study for Ham Weight Loss at the End of Dry-Curing
2.5. Candidate Gene Search for Ham Weight Loss at the End of Dry-Curing and Gene Network Analysis
3. Results and Discussion
3.1. Descriptive Statistics and Heritability for Ham Weight Loss at the End of Dry-Curing
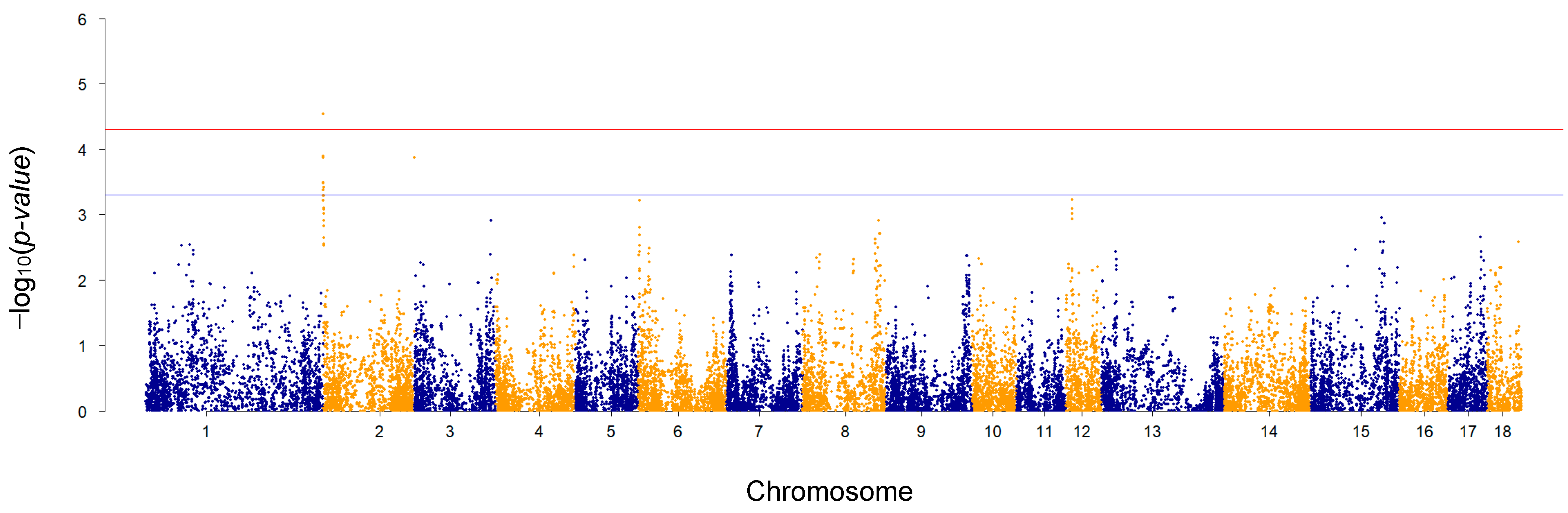
3.2. Genome-Wide Association Study for Ham Weight Loss at the End of Dry-Curing
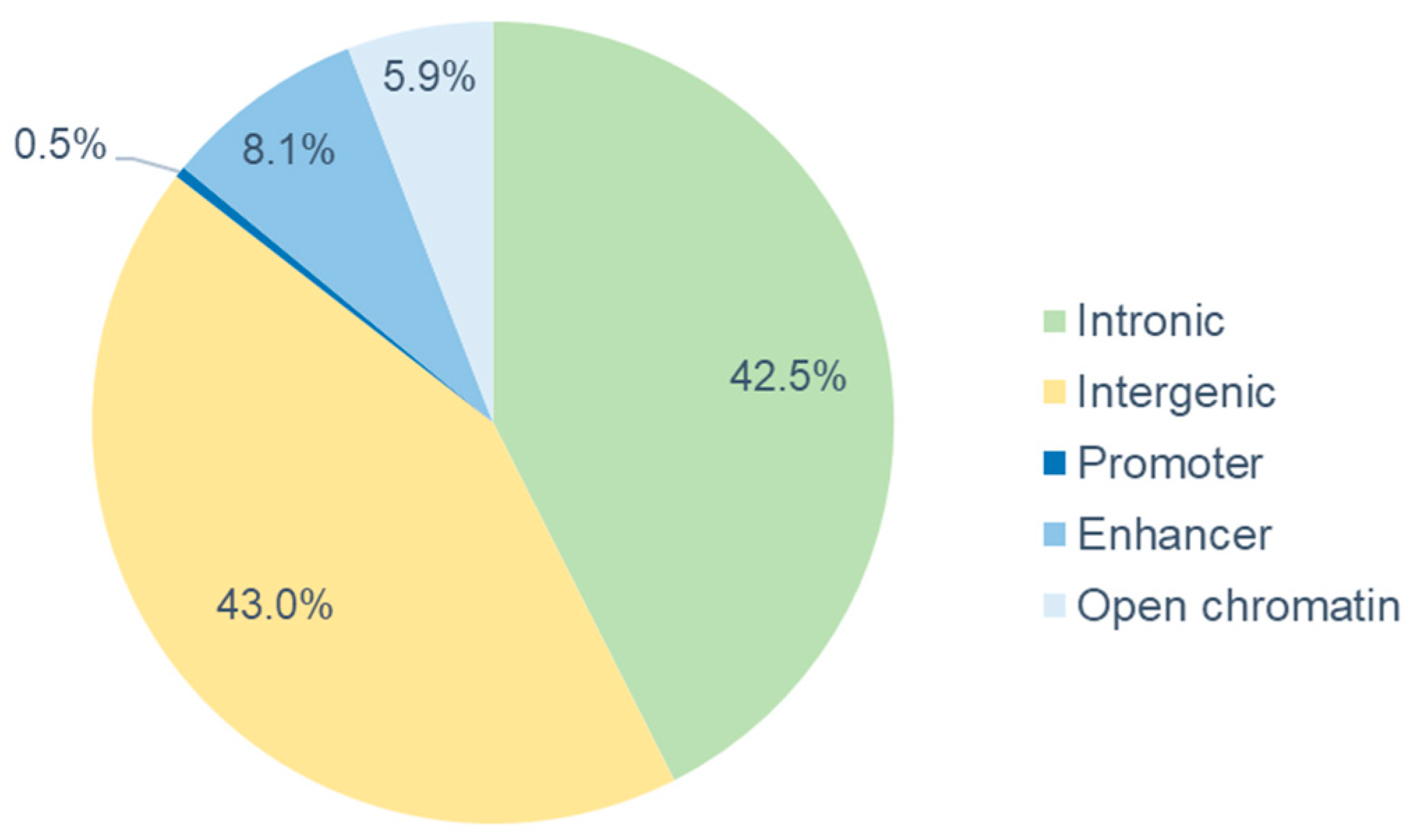
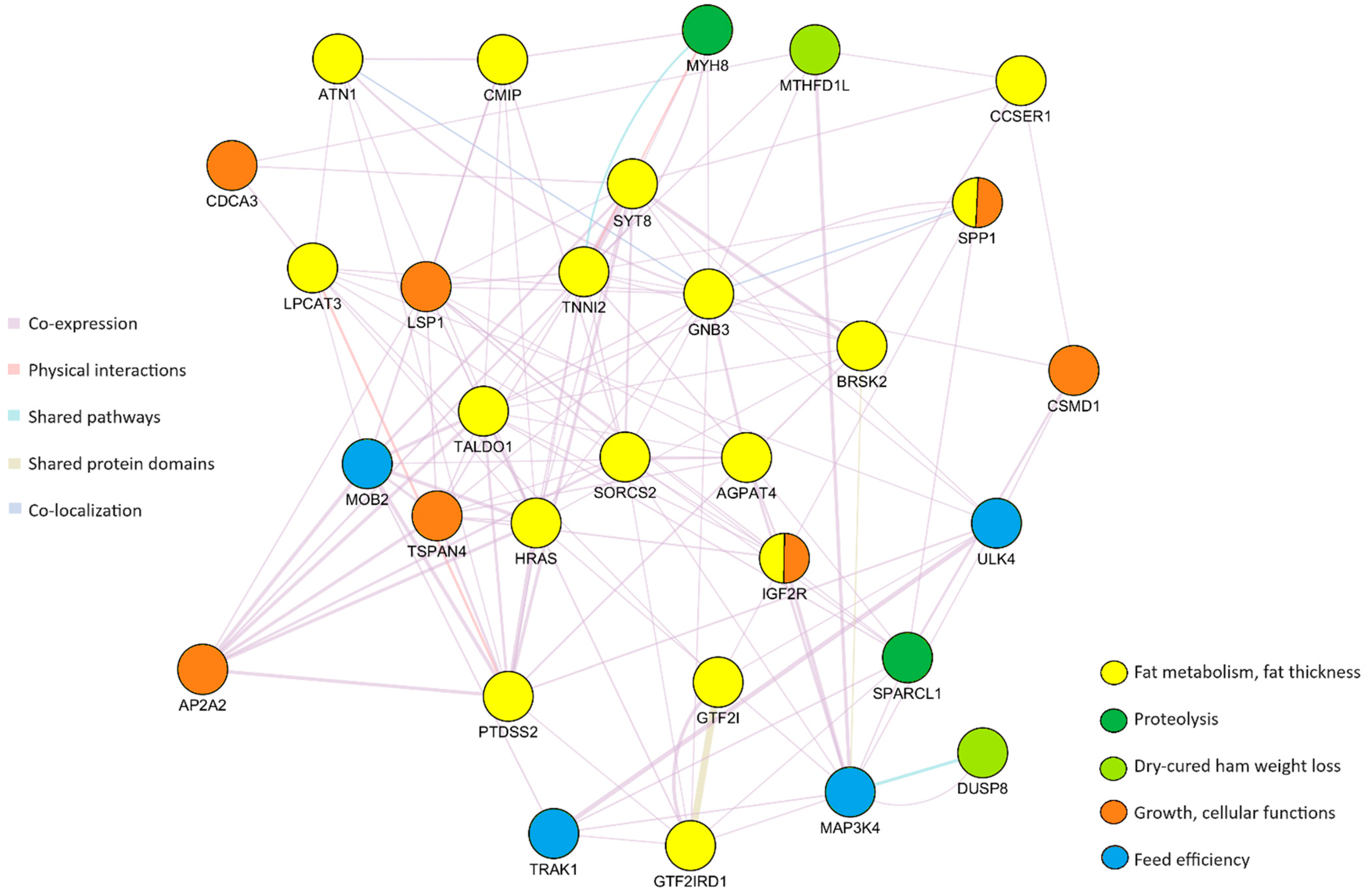
3.3. Genetic Marker Locations, Candidate Genes and Gene Network Analysis
| SSC | Position (Mb) | Gene | Name | Function | Reference |
|---|---|---|---|---|---|
| 1 | 6.875–6.875 | AGPAT4 | 1-acylglycerol-3-phosphate O-acyltransferases 4 | Fat content and composition traits; fatty acid composition | [67] |
| 1 | 6.919–6.919 | MAP3K4 | Mitogen-activated protein kinase 4 | Feed efficiency traits | [94,96] |
| 1 | 7.424–7.448 | IGF2R | Insulin-like growth factor 2 | Growth performance and carcass traits, muscle deposition, fat metabolism, meat production, and quality | [69] |
| 1 | 15.228–15.231 | MTHFD1L | Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like | Ham weight loss at first salting | [10] |
| 2 | 0.270–0.281 | PTDSS2 | Phosphatidylserine synthase 2 | Backfat thickness, fat deposition | [68] |
| 2 | 0.303–0.303 | HRAS | HRas proto-onco, GTPase | Backfat thickness at slaughter, loin depth | [9,68] |
| 2 | 0.466–0.466 | TALDO1 | Transaldolase 1 | Lipid biosynthesis | [72] |
| 2 | 0.545–0.545 | TSPAN4 | Tetraspanin 4 | Cell development, activation, growth, and motility | [71] |
| 2 | 0.633–0.633 | AP2A2 | Adaptor-related protein complex 2 subunit alpha 2 | Linking lipids in the cell membrane | [93] |
| 2 | 0.917–0.917 | BRSK2 | BR serine/threonine kinase 2 | Backfat thickness | [68] |
| 2 | 0.963–0.963 | MOB2 | MOB kinase activator 2 | Feed efficiency | [95] |
| 2 | 1.02–1.02 | DUSP8 | Dual specificity phosphatase 8 | Ham weight loss at first salting | [10] |
| 2 | 1.251–1.251 | TNNI2 | Troponin I2, fast skeletal type | Backfat thickness at slaughter; drip loss | [73,74,75] |
| 2 | 1.264–1.289 | LSP1 | Lymphocyte-specific protein 1 | Skeletal muscle development | [91] |
| 2 | 1.251–1.251 | SYT8 | Synaptotagmin 8 | Lipid metabolism | [76,77] |
| 3 | 11.578–11.643 | GTF2IRD1 | GTF2I repeat domain-containing 1 | Intramuscular fat content | [80] |
| 3 | 11.674–11.786 | GTF2I | General transcription factor IIi | Intramuscular fat content | [80] |
| 5 | 63.717–63.717 | LPCAT3 | Lysophosphatidylcholine acyltransferase 3 | Intramuscular and subcutaneous adipocytes | [70] |
| 5 | 63.794–63.794 | ATN1 | Atrophin 1 | Fat deposition in the early stages of development | [86] |
| 5 | 63.860–63.860 | GNB3 | G protein subunit beta 3 | Energy homeostasis and promotion of lipolysis | [87] |
| 5 | 63.860–63.860 | CDCA3 | Cell Division Cycle-Associated 3 | Body weight | [90] |
| 6 | 6.697–6.697 | CMIP | C-Maf-Inducing Protein | Lipid metabolism | [81] |
| 8 | 3.245–3.317 | SORCS2 | Sortilin-related VPS10 domain-containing receptor 2 | Backfat traits | [83] |
| 8 | 128.628–128.795 | CCSER1 | Coiled-coil serine rich protein 1 | Backfat thickness | [85] |
| 8 | 131.075–131.075 | SPP1 | Secreted phosphoprotein 1 | Body length, backfat thickness, loin muscle area | [89] |
| 8 | 131.392–131.410 | SPARCL1 | SPARC-like 1 | Calcium ion binding activity, proteolysis | [58] |
| 12 | 55.166–55.166 | MYH8 | Myosin heavy chain 8 | Calcium ion binding activity, proteolysis | [59] |
| 13 | 25.730–25.730 | ULK4 | Unc-51-like kinase 4 | Feed efficiency traits | [94] |
| 13 | 25.864–25.930 | TRAK1 | Trafficking kinesin protein 1 | Feed efficiency traits | [94] |
| 15 | 34.488–34.886 | CSMD1 | CUB and sushi multiple domains 1 | Cellular functions control, interaction with growth factors | [92] |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yang, Y.; Zhou, R.; Li, K. Future livestock breeding: Precision breeding based on multi-omics information and population personalization. J. Integr. Agric. 2017, 16, 2784–2791. [Google Scholar] [CrossRef]
- Ruan, D.; Zhuang, Z.; Ding, R.; Qiu, Y.; Zhou, S.; Wu, J.; Xu, C.; Hong, L.; Huang, S.; Zheng, E.; et al. Weighted single-step GWAS identified candidate genes associated with growth traits in a Duroc pig population. Genes 2021, 12, 117. [Google Scholar] [CrossRef]
- Palombo, V.; D’Andrea, M.; Licastro, D.; Dal Monego, S.; Sgorlon, S.; Sandri, M.; Stefanon, B. Single-step genome wide association study identifies QTL signals for untrimmed and trimmed thigh weight in Italian crossbred pigs for dry-cured ham production. Animals 2021, 11, 1612. [Google Scholar] [CrossRef]
- Lopez, B.I.; An, N.; Srikanth, K.; Lee, S.; Oh, J.-D.; Shin, D.-H.; Park, W.; Chai, H.-H.; Park, J.-E.; Lim, D. Genomic prediction based on SNP functional annotation using imputed whole-genome sequence data in Korean Hanwoo cattle. Front. Genet. 2021, 11, 603822. [Google Scholar] [CrossRef] [PubMed]
- Ros-Freixedes, R.; Johnsson, M.; Whalen, A.; Chen, C.-Y.; Valente, B.D.; Herring, W.O.; Gorjanc, G.; Hickey, J.M. Genomic prediction with whole-genome sequence data in intensely selected pig lines. Genet. Sel. Evol. 2022, 54, 65. [Google Scholar] [CrossRef]
- Chakraborty, D.; Sharma, N.; Kour, S.; Sodhi, S.S.; Gupta, M.K.; Lee, S.J.; Son, Y.O. Applications of OMICS technology for livestock selection and improvement. Front. Genet. 2022, 13, 774113. [Google Scholar] [CrossRef]
- Samorè, A.B.; Fontanesi, L. Genomic selection in pigs: State of the art and perspectives. Ital. J. Anim. Sci. 2016, 15, 211–232. [Google Scholar] [CrossRef]
- Sanchez, M.-P.; Tribout, T.; Iannuccelli, N.; Bouffaud, M.; Servin, B.; Tenghe, A.; Dehais, P.; Muller, N.; Del Schneider, M.; Mercat, M.-J.; et al. A genome-wide association study of production traits in a commercial population of large white pigs: Evidence of haplotypes affecting meat quality. Genet. Sel. Evol. 2014, 46, 12. [Google Scholar] [CrossRef]
- Sevillano, C.A.; ten Napel, J.; Guimarães, S.E.; Silva, F.F.; Calus, M.P. Effects of alleles in crossbred pigs estimated for genomic prediction depend on their breed-of-origin. BMC Genom. 2018, 19, 740. [Google Scholar] [CrossRef]
- Bergamaschi, M.; Maltecca, C.; Fix, J.; Schwab, C.; Tiezzi, F. Genome-wide association study for carcass quality traits and growth in purebred and crossbred pigs. J. Anim. Sci. 2020, 98, skz360. [Google Scholar] [CrossRef]
- Fontanesi, L.; Schiavo, G.; Gallo, M.; Baiocco, C.; Galimberti, G.; Bovo, S.; Russo, V.; Buttazzoni, L. Genome-wide association study for ham weight loss at first salting in Italian large white pigs: Towards the genetic dissection of a key trait for dry-cured ham production. Anim. Genet. 2016, 48, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, L.; Schiavo, G.; Galimberti, G.; Bovo, S.; Russo, V.; Gallo, M.; Buttazzoni, L. A genome-wide association study for a proxy of intermuscular fat level in the Italian large white breed identifies genomic regions affecting an important quality parameter for dry-cured hams. Anim. Genet. 2017, 48, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Schiavo, G.; Galimberti, G.; Bovo, S.; D’Andrea, M.; Gallo, M.; Buttazzoni, L.; Rothschild, M.F.; Fontanesi, L. Genome-wide association studies for seven production traits highlight genomic regions useful to dissect dry-cured ham quality and production traits in Duroc heavy pigs. Animal 2018, 12, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Stalder, K.J.; Moeller, S.J.; Rothschild, M.F. Associations between two gene markers and traits affecting fresh and dry-cured ham processing quality. Meat Sci. 2005, 69, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Renaville, B.; Piasentier, E.; Fan, B.; Vitale, M.; Prandi, A.; Rothschild, M.F. Candidate gene markers involved in San Daniele Ham Quality. Meat Sci. 2010, 85, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Centro Ricerche Produzioni Animali. Suinicoltura Italiana e Costo di Produzione; Brochure No. 1; Centro Ricerche Produzioni Animali: Reggio Emilia, Italy, 2010. [Google Scholar]
- ISMEA. Istituto di Servizi per il Mercato Agricolo Alimentare, Tendenze e Dinamiche Recenti—Suino; ISMEA: Rome, Italy, 2023. [Google Scholar]
- Buttazzoni, L.; Gallo, M.; Baiocco, C.; Carchedi, C. La selezione per la qualità della carne suina destinata alla trasformazione. Riv. Suinic. 1993, 34, 139–145. [Google Scholar]
- Malgwi, I.H.; Gallo, L.; Halas, V.; Bonfatti, V.; Carcò, G.; Sasso, C.P.; Carnier, P.; Schiavon, S. The implications of changing age and weight at slaughter of heavy pigs on carcass and green ham quality traits. Animals 2021, 11, 2447. [Google Scholar] [CrossRef] [PubMed]
- Schiavon, S.; Malgwi, I.H.; Giannuzzi, D.; Galassi, G.; Rapetti, L.; Carnier, P.; Halas, V.; Gallo, L. Impact of rearing strategies on the metabolizable energy and SID lysine partitioning in pigs growing from 90 to 200 kg in body weight. Animals 2022, 12, 689. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (NRC). Nutrient Requirements of Swine, 11th ed.; National Academy Press: Washington, DC, USA, 2012. [Google Scholar]
- Schiavon, S.; Bona, M.D.; Carcò, G.; Carraro, L.; Bunger, L.; Gallo, L. Effects of feed allowance and indispensable amino acid reduction on feed intake, growth performance and carcass characteristics of growing pigs. PLoS ONE 2018, 13, e0195645. [Google Scholar] [CrossRef]
- Prosciutto Veneto Berico-Euganeo. Protected Designation of Origin. Publication Pursuant to Article 4 of the Council Regulation (EEC) No. 2081/92 of July, 14th 1992. Official Journal of the European Union, 1—55. 1992. Available online: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/3339 (accessed on 28 March 2024).
- Toscano, A.; Giannuzzi, D.; Malgwi, I.H.; Halas, V.; Carnier, P.; Gallo, L.; Schiavon, S. Impact of innovative rearing strategies for the Italian heavy pigs: Technological traits and chemical composition of dry–cured hams. Meat Sci. 2023, 204, 109266. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 15 January 2024).
- Sargolzaei, M.; Chesnais, J.P.; Schenkel, F.S. A new approach for efficient genotype imputation using information from relatives. BMC Genom. 2014, 15, 478. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Tsuruta, S.; Lourenco, D.; Masuda, Y.; Aguilar, I.; Legarra, A.; Vitezica, Z. Manual for BLUPf90 Family of Programs. 2018. Available online: http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90_all7.pdf (accessed on 8 April 2024).
- VanRaden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Misztal, I.; Aguilar, I.; Legarra, A.; Muir, W.M. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet. Res. 2012, 94, 73–83. [Google Scholar] [CrossRef]
- Veroneze, R.; Bastiaansen, J.W.M.; Knol, E.F.; Guimarães, S.E.F.; Silva, F.F.; Harlizius, B.; Lopes, M.S.; Lopes, P.S. Linkage disequilibrium patterns and persistence of phase in purebred and crossbred pig (Sus scrofa) populations. BMC Genet. 2014, 15, 126. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, I.; Legarra, A.; Cardoso, F.; Masuda, Y.; Lourenco, D.; Misztal, I. Frequentist P-values for large-scale-single step genome-wide association, with an application to birth weight in American Angus cattle. Genet. Sel. Evol. 2019, 51, 28. [Google Scholar] [CrossRef] [PubMed]
- Gualdrón Duarte, J.L.; Cantet, R.J.C.; Bates, R.O.; Ernst, C.W.; Raney, N.E.; Steibel, J.P. Rapid screening for phenotype-genotype associations by linear transformations of genomic evaluations. BMC Bioinform. 2014, 15, 246. [Google Scholar] [CrossRef]
- Fontanesi, L.; Schiavo, G.; Galimberti, G.; Calò, D.G.; Scotti, E.; Martelli, P.L.; Buttazzoni, L.; Casadio, R.; Russo, V. A genome wide association study for backfat thickness in Italian Large White pigs highlights new regions affecting fat deposition including neuronal genes. BMC Genom. 2012, 13, 583. [Google Scholar] [CrossRef]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007, 447, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Turner, S.D. qqman: An R package for visualizing GWAS results using Q-Q and Manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Sollero, B.P.; Junqueira, V.S.; Gomes, C.C.; Caetano, A.R.; Cardoso, F.F. Tag SNP selection for prediction of tick resistance in Brazilian Braford and Hereford cattle breeds using Bayesian methods. Genet. Sel. Evol. 2017, 49, 49. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMania prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bosi, P.; Russo, V. The production of the heavy pig for high quality processed products. Ital. J. Anim. Sci. 2004, 3, 309–321. [Google Scholar] [CrossRef]
- Bonfatti, V.; Carnier, P. Prediction of dry-cured ham weight loss and prospects of use in a pig breeding program. Animal 2020, 14, 1128–1138. [Google Scholar] [CrossRef]
- Bonfatti, V.; Rostellato, R.; Carnier, P. Estimation of additive and dominance genetic effects on body weight, carcass and ham quality traits in heavy pigs. Animals 2021, 11, 481. [Google Scholar] [CrossRef]
- Bonfatti, V.; Faggion, S.; Boschi, E.; Carnier, P. Infrared predictions are a valuable alternative to actual measures of dry-cured ham weight loss in the training of genome-enabled prediction models. Animals 2022, 12, 814. [Google Scholar] [CrossRef] [PubMed]
- Carnier, P.; Cassandro, M.; Knol, E.; Padoan, D. Genetic parameters for some carcass and fresh ham traits of crossbred Goland pigs. In Recent Progress in Animal Science; Piva, G., Bertoni, G., Masoero, F., Bani, P., Calamari, L., Eds.; Franco Angeli Edizioni: Milano, Italy, 1999; pp. 221–223. [Google Scholar]
- Ehret, G.B. Genome-wide association studies: Contribution of Genomics to understanding blood pressure and essential hypertension. Curr. Hypertens. Rep. 2010, 12, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Devlin, B.; Roeder, K. Genomic control for association studies. Biometrics 1999, 55, 997–1004. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Hinrichs, A.L.; Larkin, E.K.; Suarez, B.K. Population stratification and patterns of linkage disequilibrium. Genet. Epidemiol. 2009, 33, S88–S92. [Google Scholar] [CrossRef]
- Lemos, M.V.; Chiaia, H.L.; Berton, M.P.; Feitosa, F.L.; Aboujaoud, C.; Camargo, G.M.; Baldi, F. Genome-wide association between single nucleotide polymorphisms with beef fatty acid profile in Nellore cattle using the single step procedure. BMC Genom. 2016, 17, 213. [Google Scholar] [CrossRef]
- Marques, D.B.; Bastiaansen, J.W.; Broekhuijse, M.L.; Lopes, M.S.; Knol, E.F.; Harlizius, B.; Guimarães, S.E.; Silva, F.F.; Lopes, P.S. Weighted single-step GWAS and gene network analysis reveal new candidate genes for semen traits in pigs. Genet. Sel. Evol. 2018, 50, 40. [Google Scholar] [CrossRef] [PubMed]
- Botelho, M.E.; Lopes, M.S.; Mathur, P.K.; Knol, E.F.; Fonseca e Silva, F.; Lopes, P.S.; Gimarães, S.E.; Marques, D.B.D.; Veroneze, R. Weighted genome-wide association study reveals new candidate genes related to boar taint compounds. Livest. Sci. 2022, 257, 104845. [Google Scholar] [CrossRef]
- John, S.; Sabo, P.J.; Thurman, R.E.; Sung, M.-H.; Biddie, S.C.; Johnson, T.A.; Hager, G.L.; Stamatoyannopoulos, J.A. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 2011, 43, 264–268. [Google Scholar] [CrossRef]
- Pennacchio, L.A.; Bickmore, W.; Dean, A.; Nobrega, M.A.; Bejerano, G. Enhancers: Five essential questions. Nat. Rev. Genet. 2013, 14, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Lenhard, B.; Sandelin, A.; Carninci, P. Metazoan promoters: Emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012, 13, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Grosschedl, R.; Birnstiel, M.L. Spacer DNA sequences upstream of the T-A-T-A-A-A-T-A sequence are essential for promotion of H2A histone gene transcription in vivo. Proc. Natl. Acad. Sci. USA 1980, 77, 7102–7106. [Google Scholar] [CrossRef]
- Furlong, E.E.; Levine, M. Developmental enhancers and chromosome topology. Science 2018, 361, 1341–1345. [Google Scholar] [CrossRef] [PubMed]
- Girardini, K.N.; Olthof, A.M.; Kanadia, R.N. Introns: The “dark matter” of the eukaryotic genome. Front. Genet. 2023, 14, 1150212. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Hou, L.; Yin, Y.; Wang, B.; Liu, C.; Zhou, W.; Niu, P.; Li, Q.; Huang, R.; Li, P. Genome-wide association study reveals new QTL and functional candidate genes for the number of ribs and carcass length in pigs. Anim. Genet. 2023, 54, 435–445. [Google Scholar] [CrossRef]
- Kawasaki, K.; Weiss, K.M. Evolutionary genetics of vertebrate tissue mineralization: The origin and evolution of the secretory calcium-binding phosphoprotein family. J. Exp. Zool. B Mol. Dev. Evol. 2005, 306, 295–316. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Barroso, M.Á.; Caraballo, C.; Silió, L.; Rodríguez, C.; Nuñez, Y.; Sánchez-Esquiliche, F.; Matos, G.; García-Casco, J.M.; Muñoz, M. Differences in the loin tenderness of Iberian pigs explained through dissimilarities in their transcriptome expression profile. Animals 2020, 10, 1715. [Google Scholar] [CrossRef]
- Toldrá, F.; Flores, M. The role of muscle proteases and lipases in flavor development during the processing of dry-cured ham. Crit. Rev. Food Sci. Nutr. 1998, 38, 331–352. [Google Scholar] [CrossRef] [PubMed]
- Čandek-Potokar, M.; Škrlep, M. Factors in pig production that impact the quality of dry-cured ham: A review. Animal 2012, 6, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Melody, J.L.; Lonergan, S.M.; Rowe, L.J.; Huiatt, T.W.; Mayes, M.S.; Huff-Lonergan, E. Early post-mortem biochemical factors influence tenderness and water-holding capacity of three porcine muscles. J. Anim. Sci. 2004, 82, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.G.; Lonergan, S.M.; Gardner, M.A.; Huff-Lonergan, E. Contribution of postmortem changes of integrin, desmin and μ-calpain to variation in water holding capacity of pork. Meat Sci. 2006, 74, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Lawson, M.A. The role of integrin degradation in post-mortem drip loss in pork. Meat Sci. 2004, 68, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Sturaro, E. Caratteristiche Tecnologiche e Qualitative di Cosce Suine Fresche Destinate Alla Trasformazione in Prodotti Tipici Stagionati: Aspetti Genetici e Relazioni con la Qualità del Prodotto Finale. Ph.D. Thesis, University of Padua, Padua, Italy, 2004. [Google Scholar]
- Ministerial Decree 5/12/2019, n. 12390. Requisiti di conformità del tipo genetico impiegato per la riproduzione dei suini utilizzati nel circuito delle DOP. Available online: https://www.politicheagricole.it/flex/cm/pages/ServeBLOB.php/L/IT/IDPagina/14781#:~:text=D.M.%20N.-,12390%20del%2005%2F12%2F2019%20%2D%20Requisiti%20di%20conformit%C3%A0%20del,utilizzati%20nel%20circuito%20delle%20DOP (accessed on 18 April 2024).
- Molinero, E.; Pena, R.N.; Estany, J.; Ros-Freixedes, R. Identification of a missense variant in the porcine AGPAT gene family associated with intramuscular fat content through whole-genome sequencing. Anim. Genet. 2022, 53, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Gozalo-Marcilla, M.; Buntjer, J.; Johnsson, M.; Batista, L.; Diez, F.; Werner, C.R.; Chen, C.-Y.; Gorjanc, G.; Mellanby, R.J.; Hickey, J.M.; et al. Genetic architecture and major genes for backfat thickness in pig lines of diverse genetic backgrounds. Genet. Sel. Evol. 2021, 53, 76. [Google Scholar] [CrossRef]
- Duo, T.; Liu, X.; Mo, D.; Bian, Y.; Cai, S.; Wang, M.; Li, R.; Zhu, Q.; Tong, X.; Liang, Z.; et al. Single-base editing in IGF2 improves meat production and intramuscular fat deposition in Liang Guang small spotted pigs. J. Anim. Sci. Biotechnol. 2023, 14, 141. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.; Chen, W.; Li, J.; Shan, T. CRTC3 regulates the lipid metabolism and adipogenic differentiation of porcine intramuscular and subcutaneous adipocytes by activating the calcium pathway. J. Agric. Food Chem. 2021, 69, 7243–7255. [Google Scholar] [CrossRef]
- Kwak, W.; Kim, J.; Kim, D.; Hong, J.S.; Jeong, J.H.; Kim, H.; Cho, S.; Kim, Y.Y. Genome-wide DNA methylation profiles of small intestine and liver in fast-growing and slow-growing weaning piglets. Asian-Australas. J. Anim. Sci. 2014, 27, 1532–1539. [Google Scholar] [CrossRef]
- Moriyama, T.; Tanaka, S.; Nakayama, Y.; Fukumoto, M.; Tsujimura, K.; Yamada, K.; Bamba, T.; Yoneda, Y.; Fukusaki, E.; Oka, M. Two isoforms of TALDO1 generated by alternative translational initiation show differential nucleocytoplasmic distribution to regulate the global metabolic network. Sci. Rep. 2016, 6, 34648. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Yang, H.; Li, Y.; Xiong, Y.Z.; Zuo, B. Temporal expression of TnI fast and slow isoforms in biceps femoris and masseter muscle during pig growth. Animal 2010, 4, 1541–1546. [Google Scholar] [CrossRef]
- Yang, H.; Xu, Z.Y.; Lei, M.G.; Li, F.E.; Deng, C.Y.; Xiong, Y.Z.; Zuo, B. Association of 3 polymorphisms in porcine troponin I genes (TNNI1 and TNNI2) with meat quality traits. J. Appl. Genet. 2010, 51, 51–57. [Google Scholar] [CrossRef]
- Ngu, N.; Nhan, N. Analysis of troponin I gene polymorphisms and meat quality in Mongcai pigs. S. Afr. J. Anim. Sci. 2012, 42, 288–295. [Google Scholar] [CrossRef][Green Version]
- Xu, Z.; Wei, G.; Chepelev, I.; Zhao, K.; Felsenfeld, G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat. Struct. Mol. Biol. 2011, 18, 372–378. [Google Scholar] [CrossRef]
- Moghadam, P.K.; Jackson, M.B. The functional significance of synaptotagmin diversity in neuroendocrine secretion. Front. Endocrinol. 2013, 4, 124. [Google Scholar] [CrossRef]
- Palmer, S.J.; Tay, E.S.E.; Santucci, N.; Cuc Bach, T.T.; Hook, J.; Lemckert, F.A.; Jamieson, R.V.; Gunnning, P.W.; Hardeman, E.C. Expression of GTF2IRD1, the Williams syndrome-associated gene, during mouse development. Gene Expr. Patterns 2007, 7, 396–404. [Google Scholar] [CrossRef]
- Mera, L. GTF2IRD1 Is a PRDM16-Interacting Transcription Factor That Represses TGF-β-Mediated Inhibition of Beige Fat Differentiation; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Ding, R.; Yang, M.; Quan, J.; Li, S.; Zhuang, Z.; Zhou, S.; Zheng, E.; Hong, L.; Li, Z.; Cai, G.; et al. Single-locus and multi-locus genome-wide association studies for intramuscular fat in Duroc Pigs. Front. Genet. 2019, 10, 619. [Google Scholar] [CrossRef]
- González-Prendes, R.; Quintanilla, R.; Amills, M. Investigating the genetic regulation of the expression of 63 lipid metabolism genes in the pig skeletal muscle. Anim. Genet. 2017, 48, 606–610. [Google Scholar] [CrossRef]
- Subkhangulova, A.; Malik, A.R.; Hermey, G.; Popp, O.; Dittmar, G.; Rathjen, T.; Poy, M.N.; Stumpf, A.; Beed, P.S.; Schmitz, D.; et al. SORCS1 and SORCS3 control energy balance and orexigenic peptide production. EMBO Rep. 2018, 19, e44810. [Google Scholar] [CrossRef]
- Ding, R.; Zhuang, Z.; Qiu, Y.; Ruan, D.; Wu, J.; Ye, J.; Cao, L.; Zhou, S.; Zheng, E.; Huang, W.; et al. Identify known and novel candidate genes associated with backfat thickness in Duroc pigs by large-scale genome-wide association analysis. J. Anim. Sci. 2022, 100, skac012. [Google Scholar] [CrossRef]
- Júnior, G.A.; Costa, R.B.; de Camargo, G.M.; Carvalheiro, R.; Rosa, G.J.; Baldi, F.; Garcia, D.A.; Gordo, D.G.; Espigolan, R.; Takada, L.; et al. Genome scan for post-mortem carcass traits in Nellore cattle. J. Anim. Sci. 2016, 94, 4087–4095. [Google Scholar] [CrossRef]
- Xue, Y.; Li, C.; Duan, D.; Wang, M.; Han, X.; Wang, K.; Qiao, R.; Li, X.-J.; Li, X.-L. Genome-wide association studies for growth-related traits in a crossbreed pig population. Anim. Genet. 2020, 52, 217–222. [Google Scholar] [CrossRef]
- Serão, N.V.L.; Veroneze, R.; Ribeiro, A.M.F.; Verardo, L.L.; Braccini Neto, J.; Gasparino, E.; Campos, C.F.; Lopes, P.S.; Guimarães, S.E.F. Candidate gene expression and intramuscular fat content in pigs. J. Anim. Breed. Genet. 2010, 128, 28–34. [Google Scholar] [CrossRef]
- Nowacka-Woszuk, J.; Szczerbal, I.; Fijak-Nowak, H.; Switonski, M. Chromosomal localization of 13 candidate genes for human obesity in the pig genome. J. Appl. Genet. 2008, 49, 373–377. [Google Scholar] [CrossRef]
- Yoshitake, H.; Rittling, S.R.; Denhardt, D.T.; Noda, M. Osteopontin-deficient mice are resistant to ovariectomy-induced bone resorption. Proc. Natl. Acad. Sci. USA 1999, 96, 8156–8160. [Google Scholar] [CrossRef]
- Han, S.-H.; Shin, K.-Y.; Lee, S.-S.; Ko, M.-S.; Oh, H.-S.; Cho, I.-C. Porcine SPP1 gene polymorphism association with phenotypic traits in the Landrace × Jeju (Korea) black pig F2 population. Mol. Biol. Rep. 2012, 39, 7705–7709. [Google Scholar] [CrossRef]
- Vahedi, S.M.; Salek Ardestani, S.; Karimi, K.; Banabazi, M.H. Weighted single-step GWAS for body mass index and scans for recent signatures of selection in Yorkshire pigs. J. Hered. 2022, 113, 325–335. [Google Scholar] [CrossRef]
- Albuquerque, A.; Óvilo, C.; Núñez, Y.; Benítez, R.; López-Garcia, A.; García, F.; do Félix, M.; Laranjo, M.; Charneca, R.; Martins, J.M. Transcriptomic profiling of skeletal muscle reveals candidate genes influencing muscle growth and associated lipid composition in Portuguese local pig breeds. Animals 2021, 11, 1423. [Google Scholar] [CrossRef]
- Akyuz, E.; Bell, S.M. The diverse role of CUB and sushi multiple domains 1 (CSMD1) in human diseases. Genes 2022, 13, 2332. [Google Scholar] [CrossRef]
- Kim, S.-S.; Kim, S.-R.; Kim, J.-R.; Moon, J.-K.; Choi, B.-H.; Lee, J.-W.; Kim, K.-S.; Kim, T.-H.; Kim, H.-J.; Lee, C.-K. Differences in hepatic gene expression as a major distinguishing factor between Korean native pig and Yorkshire. Biosci. Biotechnol. Biochem. 2011, 75, 451–458. [Google Scholar] [CrossRef]
- Sahana, G.; Kadlecová, V.; Hornshøj, H.; Nielsen, B.; Christensen, O.F. A genome-wide association scan in pig identifies novel regions associated with feed efficiency trait. J. Anim. Sci. 2013, 91, 1041–1050. [Google Scholar] [CrossRef]
- Mota, L.F.; Santos, S.W.; Júnior, G.A.; Bresolin, T.; Mercadante, M.E.; Silva, J.A.; Cyrillo, J.N.; Monteiro, F.M.; Carvalheiro, R.; Albuquerque, L.G. Meta-analysis across Nellore cattle populations identifies common metabolic mechanisms that regulate feed efficiency-related traits. BMC Genom. 2022, 23, 424. [Google Scholar] [CrossRef]
- Banerjee, P.; Okstoft Carmelo, V.A.; Kadarmideen, H.N. Genome-wide epistatic interaction networks affecting feed efficiency in Duroc and Landrace pigs. Front. Genet. 2020, 11, 121. [Google Scholar] [CrossRef]
- Cai, W.; Casey, D.S.; Dekkers, J.C. Selection response and genetic parameters for residual feed intake in Yorkshire swine. J. Anim. Sci. 2008, 86, 287–298. [Google Scholar] [CrossRef]
- Hsu, W.-L.; Young, J.; Mauch, E.D.; Dekkers, J.C. Analysis of ten generations of selection for residual feed intake in Yorkshire pigs. Iowa State Univ. Anim. Ind. Rep. 2015, 12, R3032. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faggion, S.; Bonfatti, V.; Carnier, P. Genome-Wide Association Study for Weight Loss at the End of Dry-Curing of Hams Produced from Purebred Heavy Pigs. Animals 2024, 14, 1983. https://doi.org/10.3390/ani14131983
Faggion S, Bonfatti V, Carnier P. Genome-Wide Association Study for Weight Loss at the End of Dry-Curing of Hams Produced from Purebred Heavy Pigs. Animals. 2024; 14(13):1983. https://doi.org/10.3390/ani14131983
Chicago/Turabian StyleFaggion, Sara, Valentina Bonfatti, and Paolo Carnier. 2024. "Genome-Wide Association Study for Weight Loss at the End of Dry-Curing of Hams Produced from Purebred Heavy Pigs" Animals 14, no. 13: 1983. https://doi.org/10.3390/ani14131983
APA StyleFaggion, S., Bonfatti, V., & Carnier, P. (2024). Genome-Wide Association Study for Weight Loss at the End of Dry-Curing of Hams Produced from Purebred Heavy Pigs. Animals, 14(13), 1983. https://doi.org/10.3390/ani14131983






