Comparative Transcriptome Analysis of mRNA and miRNA during the Development of Longissimus Dorsi Muscle of Gannan Yak and Tianzhu White Yak
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Laboratory Animals and Sample Collection
2.2. Analysis of Muscle Fiber Characteristics
2.3. Total RNA Extraction and Sequencing
2.4. Data Quality Control and Differential miRNA Screening and Identification
2.5. (DE) miRNA Target Gene Prediction and Functional Enrichment Analysis
2.6. (DE)miRNA-(DE)mRNA Regulatory Network Analysis
2.7. Validation of qRT-PCR for (DE) miRNAs
3. Results
3.1. Analysis of Dyeing Characteristics of Muscle Fibers
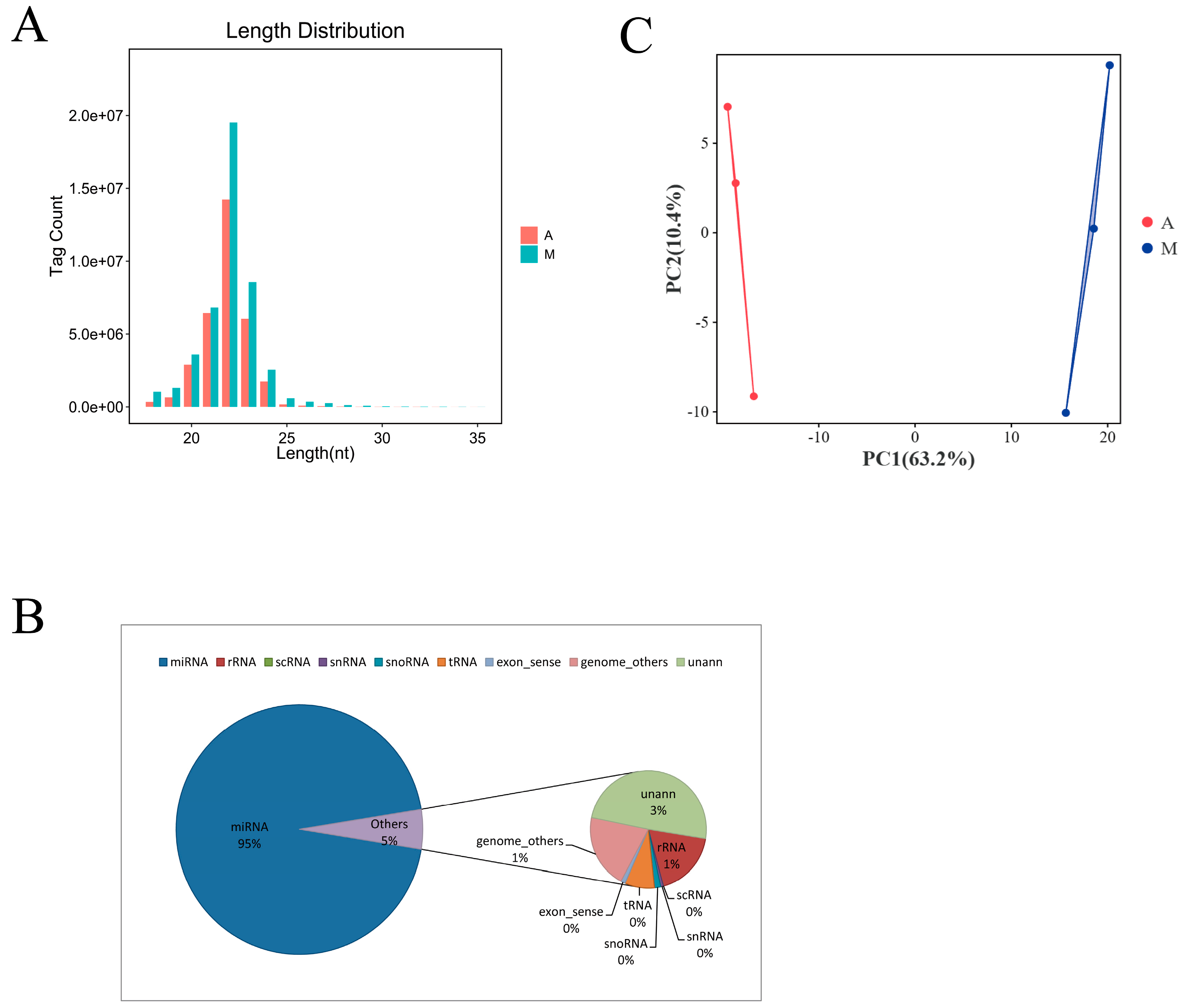
3.2. Identification and Characterization of miRNAs
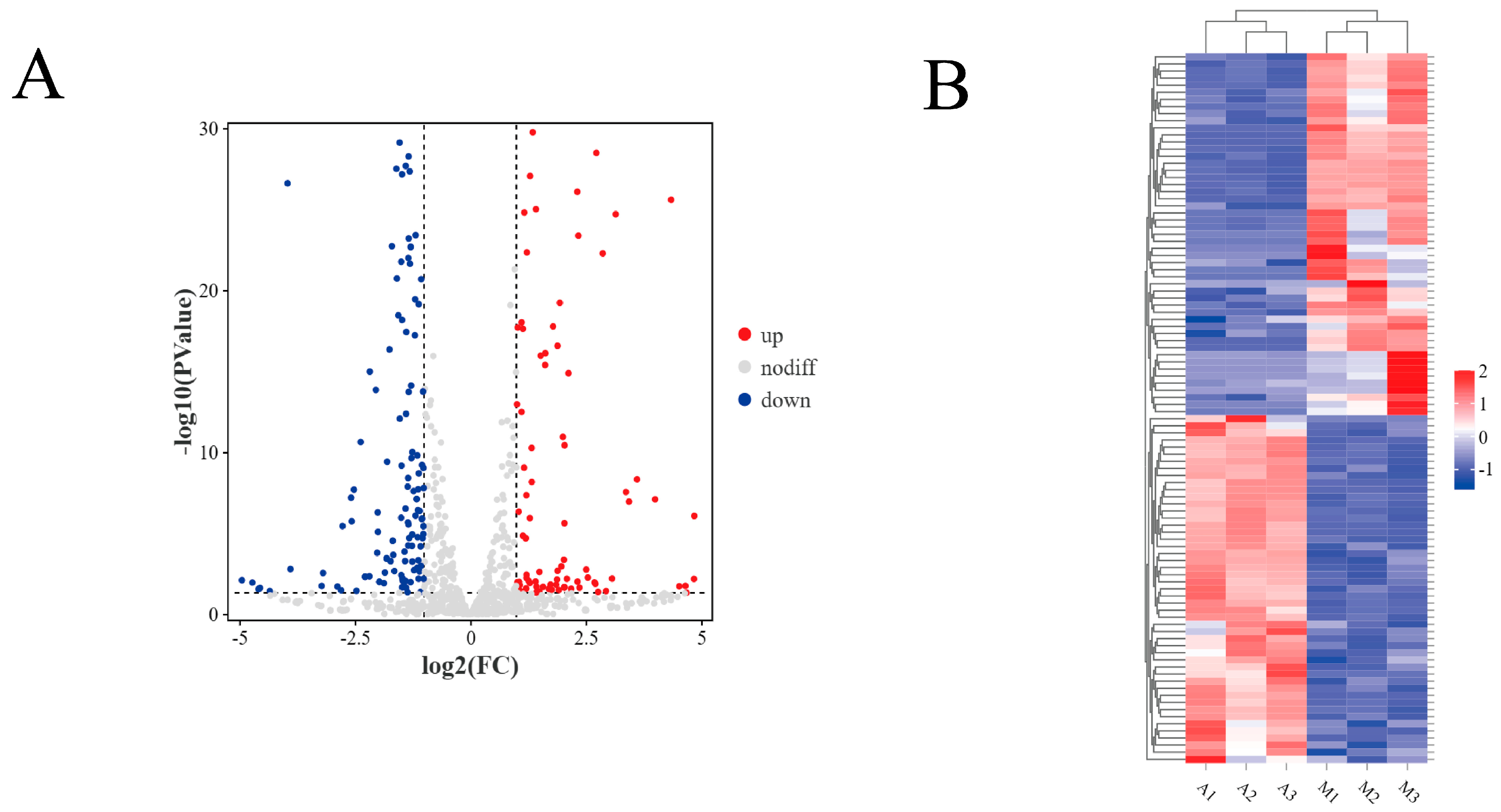
3.3. Differential Expression Analysis of miRNAs
3.4. Differential Expression miRNA GO and KEGG Analysis and Target Gene Prediction
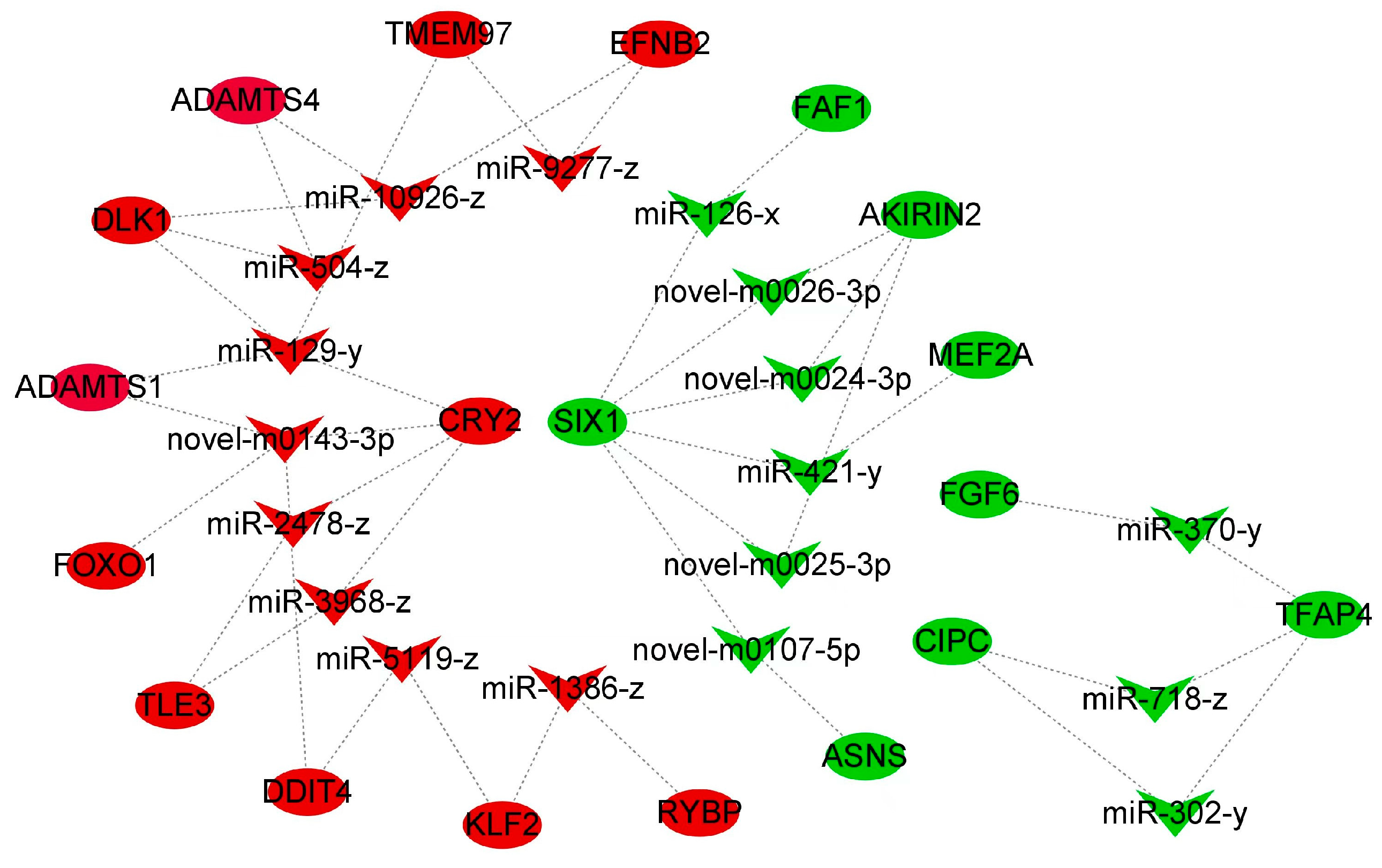
3.5. Constructing miRNA-mRNA Interaction Networks
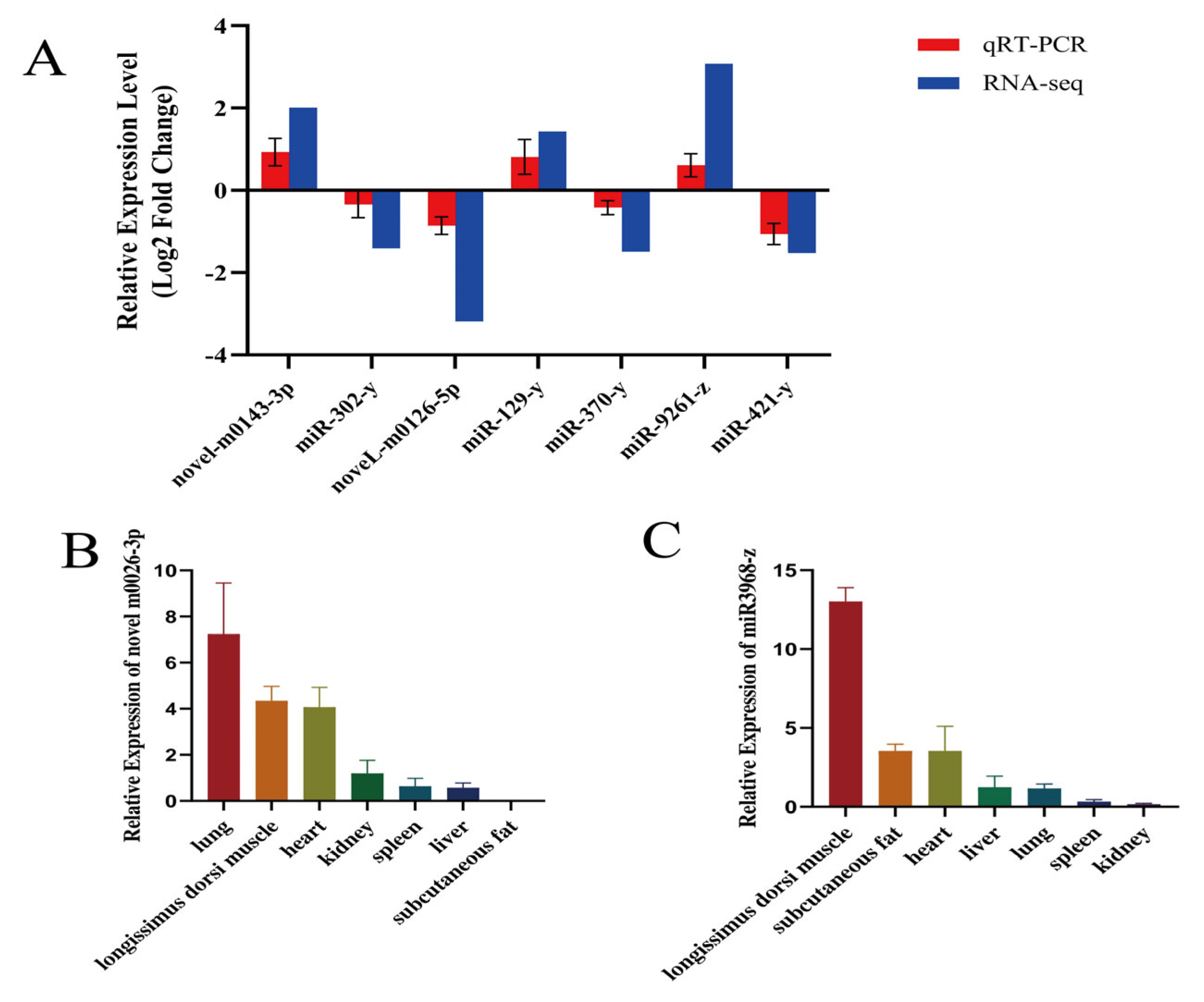
3.6. qRT-PCR Validation of Differentially Expressed miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, L.; Wei, J.; Li, J.; Wang, Y. Progress of Research on Yak Germplasm Resources and Yak Meat Quality Regulation in China. J. Gansu Agric. Univ. 2023, 58, 1–8. [Google Scholar]
- He, M. Tianzhu White Yak Development Status, Problems and Suggestions. Chin. Herbiv. 2008, 28, 59–60. [Google Scholar]
- Zhang, T.; Yang, Q.; Ma, G.; Liu, H.; Zhang, H. Conservation, Development and Utilization of Yak Germplasm Resources in Gannan. J. Anim. Husb. Vet. Med. 2016, 35, 78–79. [Google Scholar]
- Wang, Z.; Li, J.; Ma, D. Analyzing the Key Links of Gannan Yak Industry Based on Quality and Efficiency Improvement. J. Anim. Husb. Vet. Med. 2022, 41, 160–163. [Google Scholar]
- Yang, X.; Li, J.; Pan, H.; Tang, Z. Determination of some physiological indexes in Tianzhu white yak. China Yak 1987, 1, 8–14. [Google Scholar]
- Yin, H.; He, H.; Cao, X.; Shen, X.; Han, S.; Cui, C.; Zhao, J.; Wei, Y.; Chen, Y.; Xia, L.; et al. MiR-148a-3p Regulates Skeletal Muscle Satellite Cell Differentiation and Apoptosis via the PI3K/AKT Signaling Pathway by Targeting Meox2. Front. Genet. 2020, 11, 512. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Hu, X.; Cao, D.; Liu, W.; Han, H.; Zhou, Y.; Lei, Q. Deciphering the miRNA transcriptome of breast muscle from the embryonic to post-hatching periods in chickens. BMC Genom. 2021, 22, 64. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Hernandez, J.M.; Garcia-Gonzalez, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 2017, 72, 33–44. [Google Scholar] [CrossRef]
- Wei, Y.; Guo, D.; Bai, Y.; Liu, Z.; Li, J.; Chen, Z.; Shi, B.; Zhao, Z.; Hu, J.; Han, X.; et al. Transcriptome Analysis of mRNA and lncRNA Related to Muscle Growth and Development in Gannan Yak and Jeryak. Int. J. Mol. Sci. 2023, 24, 16991. [Google Scholar] [CrossRef]
- Guo, D.; Wei, Y.; Li, X.; Bai, Y.; Liu, Z.; Li, J.; Chen, Z.; Shi, B.; Zhang, X.; Zhao, Z.; et al. Comprehensive Analysis of miRNA and mRNA Expression Profiles during Muscle Development of the Longissimus Dorsi Muscle in Gannan Yaks and Jeryaks. Genes 2023, 14, 2220. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.M. The Role of AMPK in the Regulation of Skeletal Muscle Size, Hypertrophy, and Regeneration. Int. J. Mol. Sci. 2018, 19, 3125. [Google Scholar] [CrossRef] [PubMed]
- Ryall, J.G.; Church, J.E.; Lynch, G.S. Novel role for ss-adrenergic signalling in skeletal muscle growth, development and regeneration. Clin. Exp. Pharmacol. Physiol. 2010, 37, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Huang, R.; Wu, Q.; Li, P.; Chen, J.; Li, B.; Liu, H. The role of Six1 in the genesis of muscle cell and skeletal muscle development. Int. J. Biol. Sci. 2014, 10, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Rahman, F.U.; Kim, K.S.; Kim, E.K.; Cho, S.M.; Lee, K.; Moon, O.S.; Seo, Y.W.; Yoon, W.K.; Won, Y.S.; et al. Critical Roles of E2F3 in Growth and Musculo-skeletal Phenotype in Mice. Int. J. Med. Sci. 2019, 16, 1557–1563. [Google Scholar] [CrossRef] [PubMed]
- Heijmans, B.T.; Kremer, D.; Tobi, E.W.; Boomsma, D.I.; Slagboom, P.E. Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum. Mol. Genet. 2007, 16, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, C.A.; Naya, F.J. The Function of the MEF2 Family of Transcription Factors in Cardiac Development, Cardiogenomics, and Direct Reprogramming. J. Cardiovasc. Dev. Dis. 2016, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Correia, D.S.M.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; Souza, J.D.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. Camb. Philos. Soc. 2018, 93, 1955–1986. [Google Scholar] [CrossRef]
- Jia, B.; Liu, Y.; Li, Q.; Zhang, J.; Ge, C.; Wang, G.; Chen, G.; Liu, D.; Yang, F. Altered miRNA and mRNA Expression in Sika Deer Skeletal Muscle with Age. Genes 2020, 11, 172. [Google Scholar] [CrossRef]
- Deng, Z.; Chen, J.F.; Wang, D.Z. Transgenic overexpression of miR-133a in skeletal muscle. BMC Musculoskelet. Disord. 2011, 12, 115. [Google Scholar] [CrossRef] [PubMed]
- Bjorkman, K.K.; Guess, M.G.; Harrison, B.C.; Polmear, M.M.; Peter, A.K.; Leinwand, L.A. miR-206 enforces a slow muscle phenotype. J. Cell Sci. 2020, 133, jcs243162. [Google Scholar] [CrossRef] [PubMed]
- Winbanks, C.E.; Beyer, C.; Hagg, A.; Qian, H.; Sepulveda, P.V.; Gregorevic, P. miR-206 represses hypertrophy of myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS ONE 2013, 8, e73589. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Li, Z.; Wang, X.; Wang, J.; Pang, W.; Yang, G.; Shen, Q.W. microRNA-151-3p regulates slow muscle gene expression by targeting ATP2a2 in skeletal muscle cells. J. Cell. Physiol. 2015, 230, 1003–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Hou, L.; Ou, J.; Xu, G.; Jiang, F.; Hu, C.; Wang, C. MiR-199b represses porcine muscle satellite cells proliferation by targeting JAG1. Gene 2019, 691, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Xu, J.; Li, H.; Ou, J.; Jiao, Y.; Hu, C.; Wang, C. MiR-34c represses muscle development by forming a regulatory loop with Notch1. Sci. Rep. 2017, 7, 9346. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Zhang, W.R.; Wang, Y.M.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. MicroRNA-128 regulates the proliferation and differentiation of bovine skeletal muscle satellite cells by repressing Sp1. Mol. Cell. Biochem. 2016, 414, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.R.; Zhang, H.N.; Wang, Y.M.; Dai, Y.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. miR-143 regulates proliferation and differentiation of bovine skeletal muscle satellite cells by targeting IGFBP5. In Vitro Cell. Dev. Biol.-Anim. 2017, 53, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, P.; Dan, X.; Kang, X.; Ma, Y.; Shi, Y. miR-377 Inhibits Proliferation and Differentiation of Bovine Skeletal Muscle Satellite Cells by Targeting FHL2. Genes 2022, 13, 947. [Google Scholar] [CrossRef]
- Shi, B.; Shi, X.; Zuo, Z.; Zhao, S.; Zhao, Z.; Wang, J.; Zhou, H.; Luo, Y.; Hu, J.; Hick, J.G. Identification of differentially expressed genes at different post-natal development stages of longissimus dorsi muscle in Tianzhu white yak. Gene. 2022, 823, 146356. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic. Acids. Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Betel, D.; Koppal, A.; Agius, P.; Sander, C.; Leslie, C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010, 11, R90. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Mishra, B.; Qin, N.; Sun, X.; Zhang, S.; Yang, J.; Xu, R. Differential Transcriptome Analysis of Early Postnatal Developing Longissimus Dorsi Muscle from Two Pig Breeds Characterized in Divergent Myofiber Traits and Fatness. Anim. Biotechnol. 2019, 30, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.D.; Jeong, J.Y.; Yang, H.S.; Hur, S.J. Differential abundance of proteome associated with intramuscular variation of meat quality in porcine longissimus thoracis et lumborum muscle. Meat Sci. 2019, 149, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Juvvuna, P.K.; Kukreti, H.; Mcfarlane, C. Mega roles of microRNAs in regulation of skeletal muscle health and disease. Front. Physiol. 2014, 5, 239. [Google Scholar] [CrossRef] [PubMed]
- Silva-Vignato, B.; Coutinho, L.L.; Cesar, A.; Poleti, M.D.; Regitano, L.; Balieiro, J. Comparative muscle transcriptome associated with carcass traits of Nellore cattle. Bmc Genom. 2017, 18, 506. [Google Scholar] [CrossRef]
- Girardi, F.; Le Grand, F. Wnt Signaling in Skeletal Muscle Development and Regeneration. Prog. Mol. Biol. Transl. Sci. 2018, 153, 157–179. [Google Scholar]
- Cisternas, P.; Henriquez, J.P.; Brandan, E.; Inestrosa, N.C. Wnt signaling in skeletal muscle dynamics: Myogenesis, neuromuscular synapse and fibrosis. Mol. Neurobiol. 2014, 49, 574–589. [Google Scholar] [CrossRef]
- Suzuki, J.; Kaziro, Y.; Koide, H. Positive regulation of skeletal myogenesis by R-Ras. Oncogene 2000, 19, 1138–1146. [Google Scholar] [CrossRef]
- Sakakibara, I.; Wurmser, M.; Dos, S.M.; Santolini, M.; Ducommun, S.; Davaze, R.; Guernec, A.; Sakamoto, K.; Maire, P. Six1 homeoprotein drives myofiber type IIA specialization in soleus muscle. Skelet. Muscle 2016, 6, 30. [Google Scholar] [CrossRef]
- Gordon, B.S.; Delgado, D.D.; White, J.P.; Carson, J.A.; Kostek, M.C. Six1 and Six1 cofactor expression is altered during early skeletal muscle overload in mice. J. Physiol. Sci. 2012, 62, 393–401. [Google Scholar] [CrossRef]
- Chen, X.; Guo, Y.; Jia, G.; Zhao, H.; Liu, G.; Huang, Z. Effects of Active Immunization Against Akirin2 on Muscle Fiber-type Composition in Pigs. Anim. Biotechnol. 2019, 30, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luo, Y.; Huang, Z.; Jia, G.; Liu, G.; Zhao, H. Akirin2 regulates proliferation and differentiation of porcine skeletal muscle satellite cells via ERK1/2 and NFATc1 signaling pathways. Sci. Rep. 2017, 7, 45156. [Google Scholar] [CrossRef]
- Chen, X.; Luo, Y.; Huang, Z.; Liu, G.; Zhao, H. Akirin2 promotes slow myosin heavy chain expression by CaN/NFATc1 signaling in porcine skeletal muscle satellite cells. Oncotarget 2017, 8, 25158–25166. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Shih, C.H.; Wosczyna, M.N.; Mueller, A.A.; Cho, J.; Aggarwal, A.; Rando, T.A.; Feldman, B.J. Macrophage-released ADAMTS1 promotes muscle stem cell activation. Nat. Commun. 2017, 8, 669. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiao, Y.; Zheng, Y.; Yang, L.; Wang, D. An anti-ADAMTS1 treatment relieved muscle dysfunction and fibrosis in dystrophic mice. Life Sci. 2021, 281, 119756. [Google Scholar] [CrossRef]
- Hao, Y.; Xue, T.; Liu, S.B.; Geng, S.; Shi, X.; Qian, P.; He, W.; Zheng, J.; Li, Y.; Lou, J.; et al. Loss of CRY2 promotes regenerative myogenesis by enhancing PAX7 expression and satellite cell proliferation. Medcomm 2023, 4, e202. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, T.; Li, Y.; Fei, W.; Yang, G.; Hong, Y. Inhibition of CRY2 by STAT3/miRNA-7-5p Promotes Osteoblast Differentiation through Upregulation of CLOCK/BMAL1/P300 Expression. Mol. Ther. Nucleic Acids 2020, 19, 865–876. [Google Scholar] [CrossRef]


| miRNAs | Forward (5′ → 3′) | Reverse (5′ → 3′) |
|---|---|---|
| miR-129-y | AAGCCCTTACCCCAAAAAGGTAT | |
| novel-m0143-3p | CCACCAGGCCTGCAGCTCCGCC | |
| miR-9261-z | GCGGTGGGGCGCGGGACA | |
| miR-421-y | ATCAACAGACATTAATTGGGCGC | |
| novel-m0126-5p | CACTGAGACTCGCAGAAGCG | |
| miR-370-y | GCCTGCTGGGGTGGAACCTGGT | |
| miR-302-y | TAAGTGCTTCCATGTTTTAGTAG | |
| U6 | GGAACGATACAGAGAAGATTAGC | TGGAACGCTTCACGAATTTGCG |
| ID | Clean Reads | High Quality | 3′Adapter Null | Insert Null | 5′Adapter Contaminants | PolyA | Clean Tags |
|---|---|---|---|---|---|---|---|
| A1 | 11,845,237 | 11,752,376 | 5365 (0.0457%) | 46,810 (0.3983%) | 8035 (0.0684%) | 71 (0.0006%) | 11,432,116 |
| (100%) | (99.2160%) | (96.5123%) | |||||
| A2 | 11,258,234 | 12,140,269 | 8840 (0.0728%) | 34,315 (0.2827%) | 4759 (0.0392%) | 69 (0.0006%) | 11,881,484 |
| (100%) | (99.0377%) | (96.9266%) | |||||
| A3 | 9,755,904 | 9,634,408 | 3962 (0.0411%) | 32,486 (0.3372%) | 4045 (0.0420%) | 52 (0.0005%) | 9,361,019 |
| (100%) | (98.7546%) | (95.9523%) | |||||
| M1 | 17,408,914 | 17,260,011 | 9612 (0.0557%) | 108,519 (0.6287%) | 31,981 (0.1853%) | 321 (0.0019%) | 16,255,077 |
| (100%) | (99.1447%) | (93.3721%) | |||||
| M2 | 14,015,050 | 13,899,769 | 6900 (0.0496%) | 54,327 (0.3908%) | 13,958 (0.1004%) | 162 (0.0012%) | 13,362,108 |
| (100%) | (99.1774%) | (95.3411%) | |||||
| M3 | 16,597,591 | 16,448,657 | 10,968 (0.0667%) | 83,221 (0.5059%) | 20,095 (0.1222%) | 247 (0.0015%) | 15,172,291 |
| (100%) | (99.1027%) | (91.4126%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niu, Y.; Guo, D.; Wei, Y.; Li, J.; Bai, Y.; Liu, Z.; Jia, X.; Chen, Z.; Li, L.; Shi, B.; et al. Comparative Transcriptome Analysis of mRNA and miRNA during the Development of Longissimus Dorsi Muscle of Gannan Yak and Tianzhu White Yak. Animals 2024, 14, 2278. https://doi.org/10.3390/ani14152278
Niu Y, Guo D, Wei Y, Li J, Bai Y, Liu Z, Jia X, Chen Z, Li L, Shi B, et al. Comparative Transcriptome Analysis of mRNA and miRNA during the Development of Longissimus Dorsi Muscle of Gannan Yak and Tianzhu White Yak. Animals. 2024; 14(15):2278. https://doi.org/10.3390/ani14152278
Chicago/Turabian StyleNiu, Yanmei, Dashan Guo, Yali Wei, Jingsheng Li, Yanbin Bai, Zhanxin Liu, Xue Jia, Zongchang Chen, Liang Li, Bingang Shi, and et al. 2024. "Comparative Transcriptome Analysis of mRNA and miRNA during the Development of Longissimus Dorsi Muscle of Gannan Yak and Tianzhu White Yak" Animals 14, no. 15: 2278. https://doi.org/10.3390/ani14152278





